Application of the Luedeking and Piret with delay time model in bioproductions with non-zero kinetic parameters
Keywords:
Bioproductions kinetics; Delay time; Luedeking and Piret; Mathematical modeling.Abstract
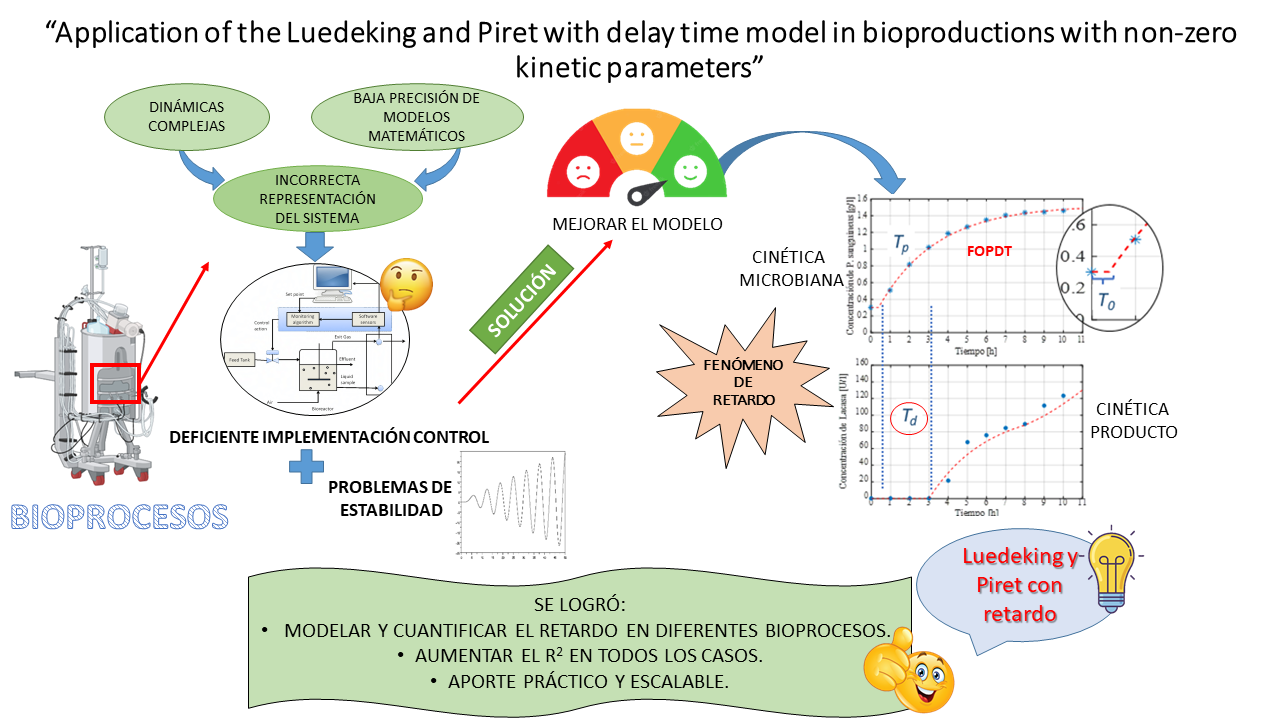
In this work, we were able to generalize the applicability of the Luedeking and Piret with delay time model previously proposed by our team. To demonstrate this, experimental data and kinetic parameters were collected from four different bioproduction. To these experimental data, the first order plus dead time (FOPDT) model was fitted for biomass production kinetics, while the Luedeking and Piret with delay time model was for different metabolites production kinetics. The use of the Luedeking and Piret with delay time model proves the delay that exists between the production time of microbial biomass and metabolites or products. The use of the FOPDT model allowed this delay to be determined in a simple and fast way. This model's combination shows an increase in R2 in all cases, demonstrating the quality of the fit and the simplicity of the proposed method. The use of delay times for bioproduction with non-zero kinetic parameters has not been previously reported. Application of this model would improve the accuracy of scaling bioprocesses to enable industrial-scale production. This delay time is an essential property of the bio-process, and its determination is crucial for control and optimization.
Downloads
References
W. World Meteorological Organization, “United in Science 2020,” 2020. [Online]. Available: https://public.wmo.int/en/resources/united_in_science
U. EPA, “Global Greenhouse Gas Emissions Data | US EPA,” 2022. https://www.epa.gov/ghgemissions/global-greenhouse-gas-emissions-data#Sector (accessed Apr. 07, 2022).
M. M. Bugge, T. Hansen, and A. Klitkou, “What Is the Bioeconomy ? A Review of the Literature,” Sustainability, vol. 8, no. 7, p. 691, 2016, doi: 10.3390/su8070691.
A. L. Demain, “The business of biotechnology,” Ind. Biotechnol., vol. 3, no. 3, pp. 269–283, 2007, doi: 10.1089/ind.2007.3.269.
C. Fernandez, N. Pantano, L. Rodriguez, and G. Scaglia, “Additive Uncertainty Consideration for Nonlinear and Multivariable Bioprocess Control,” IEEE Lat. Am. Trans., vol. 19, no. 5, pp. 798–806, 2021, doi: 10.1109/TLA.2021.9448314.
R. Luedeking and E. L. Piret, “A kinetic study of the lactic acid fermentation. Batch process at controlled pH,” J. Biochem. Microbiol. Technol. Eng., vol. 1, no. 4, pp. 393–412, 1959, doi: 10.1002/(SICI)1097-0290(20000320)67:6<636::AID-BIT3>3.0.CO;2-U.
M. C. Groff, G. Scaglia, O. A. Ortiz, and S. E. Noriega, “Modification of the Luedeking and Piret model with a delay time parameter for biotechnological lactic acid production,” Biotechnol. Lett., vol. 0, 2022, doi: 10.1007/s10529-022-03227-0.
M. F. Sardella, M. E. Serrano, O. Camacho, and G. J. E. Scaglia, “Design and Application of a Linear Algebra Based Controller from a Reduced-Order Model for Regulation and Tracking of Chemical Processes under Uncertainties,” Ind. Eng. Chem. Res., vol. 58, pp. 15222–15231, 2019, doi: 10.1021/acs.iecr.9b01257.
G. Scaglia, P. M. Aballay, M. E. Serrano, O. A. Ortiz, M. Jordan, and M. D. Vallejo, “Linear algebra based controller design applied to a bench-scale oenological alcoholic fermentation,” Control Eng. Pract., vol. 25, no. 1, pp. 66–74, 2014, doi: 10.1016/j.conengprac.2014.01.002.
M. Germec and I. Turhan, “Enhanced production of Aspergillus niger inulinase from sugar beet molasses and its kinetic modeling,” Biotechnol. Lett., vol. 42, no. 10, pp. 1939–1955, 2020, doi: 10.1007/s10529-020-02913-1.
M. N. Saat, M. S. M. Annuar, Z. Alias, L. T. Chuan, and Y. Chisti, “Modeling of growth and laccase production by Pycnoporus sanguineus,” Bioprocess Biosyst. Eng., vol. 37, no. 5, pp. 765–775, 2014, doi: 10.1007/s00449-013-1046-8.
X. Wang et al., “Modeling for gellan gum production by Sphingomonas paucimobilis ATCC 31461 in a simplified medium,” Appl Env. Microbiol, vol. 72, no. 5, pp. 3367–3374, 2006, doi: 10.1128/AEM.72.5.3367-3374.2006.
B. Liu, J. Hui, Y. Q. Cheng, and X. Zhang, “Extractive fermentation for enhanced production of thailandepsin A from Burkholderia thailandensis E264 using polyaromatic adsorbent resin Diaion HP-20,” J. Ind. Microbiol. Biotechnol., vol. 39, no. 5, pp. 767–776, May 2012, doi: 10.1007/s10295-011-1073-x.
F. D. H. Dalsenter, G. Viccini, M. C. Barga, D. A. Mitchell, and N. Krieger, “A mathematical model describing the effect of temperature variations on the kinetics of microbial growth in solid-state culture,” Process Biochem., vol. 40, pp. 801–807, 2005, doi: 10.1016/j.procbio.2004.02.007.
S. Saithi, J. Borg, M. Nopharatana, and A. Tongta, “Mathematical Modeling of Biomass and Enzyme Production Kinetics by Aspergillus niger in Solid-State Fermentation at Various Temperatures and Moisture Contents,” J. Microb. Biochem. Technol., vol. 08, no. 02, pp. 123–130, 2016, doi: 10.4172/1948-5948.1000274.
E. L. Gaden, “Fermentation process kinetics,” Biotechnol Bioener, vol. 67, pp. 629–635, 2000, doi: 10.1016/s0734-9750(97)87650-1.
B. Kuchen, S. A. Garay, R. M. Gil, F. Vazquez, and G. J. E. Scaglia, “Optimization of batch reactors : Application to the biocontrol of spoilage yeasts in wines,” IEEE Lat. Am. Trans., vol. 100(XXX), 2022, [Online]. Available: https://latamt.ieeer9.org/index.php/transactions/article/view/6934
C. Fernández, N. Pantano, S. Godoy, E. Serrano, and G. Scaglia, “Optimización de Parámetros Utilizando los Métodos de Monte Carlo y Algoritmos Evolutivos. Aplicación a un Controlador de Seguimiento de Trayectoria en Sistemas no Lineales,” Rev. Iberoam. Automática e Informática Ind., vol. 00, no. 2011, pp. 1–4, 2018, doi: 10.4995/ riai.2018.