Prediction of Biomethane Production of Cheese Whey by Using Artificial Neural Networks
Keywords:
Biomethane, Kinetics, Cheese whey, Modelling, Artificial neural networkAbstract
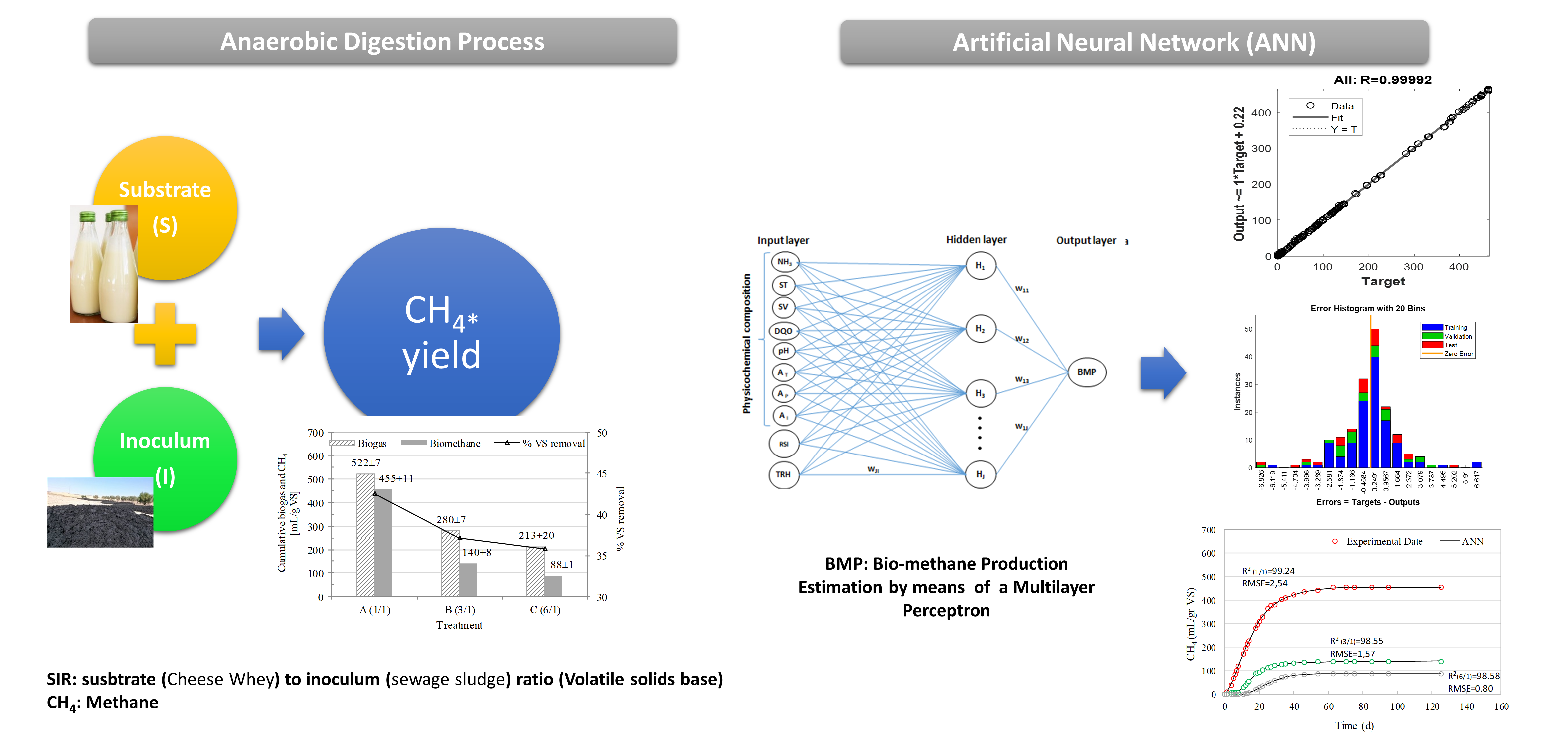
The search for new energy sources has intensified today worldwide. In Argentina, bioenergy continues to be the energy with minor participation in the energy matrix. Therefore, research should focus on searching for new substrates that increase their production without compromising agricultural systems. This work analyzed the cumulative methane production by anaerobic digestion of cheese whey, a food industry waste, using three substrate/inoculum ratios in volatile solids units. An artificial neural network (ANN) was developed to model the process based on the characterization data (input data) and methane production obtained in the laboratory (output data). The training algorithm used for the ANN was backpropagation. This model's validity was analyzed using statistical parameters such as the regression coefficient (R2) and the mean square error (MSE). The results obtained by the ANN were compared with those obtained by conventional kinetic models, such as the First-order and Gompertz models. The experimental biomethane production was in the range 232±5 to 382±22 NmLCH4/g VS. The proposed network could predict the experimental data with an R2 of 0.9992 and a training MSE of 2.7598. The statistics used to compare the goodness of fit between the models (R2 and RMSE) were higher for the network, demonstrating its ability to model a complex system such as anaerobic digestion.
Downloads
References
A. G. Nogar, L. D. Rodríguez, C.-V. Bongiorno, and E. M. Santalla, ‘Las potencialidades bioenergéticas del Arundo donax L. en Argentina’, Let. Verdes. Rev. Latinoam. Estud. Socioambientales, no. 30, pp. 84–104, 2021, doi: 10.17141/letrasverdes.30.2021.4607.
J. Liebetrau, U. Baier, D. Wall, and J. D. Murphy, Integration of biogas systems into the energy system, no. 8. IEA Bioenergy Task 37, 2020.
A. Scott and R. Blanchard, ‘The role of anaerobic digestion in reducing dairy farm greenhouse gas emissions’, Sustain., vol. 13, no. 5, pp. 1–18, 2021, doi: 10.3390/su13052612.
A. H. Geeraerd, C. H. Herremans, C. Cenens, and J. F. Van Impe, ‘Application of artificial neural networks as a non-linear modular modeling technique to describe bacterial growth in chilled food products’, Int. J. Food Microbiol., vol. 44, pp. 49–68, 1998.
R. B. Recio Colmenares and K. J. Gurubel Tún, ‘Modelado neuronal de un proceso de digestión aeróbica de aguas residuales’, Bistua Rev. La Fac. Ciencias Basicas, vol. 16, no. 1, p. 163, 2019, doi: 10.24054/01204211.v1.n1.2018.3204.
C. Holliger et al., ‘Towards a standardization of biomethane potential tests’, Water Sci. Technol., vol. 74, no. 11, pp. 1–9, 2016, doi: 10.2166/wst.2016.336.
K. Yetilmezsoy, F. I. Turkdogan, I. Temizel, and A. Gunay, ‘Development of ann-based models to predict biogas and methane productions in anaerobic treatment of molasses wastewater’, Int. J. Green Energy, vol. 10, no. 9, pp. 885–907, 2013, doi: 10.1080/15435075.2012.727116.
D. J. Batstone, J. Keller, I. Angelidaki, S. V Kalyuzhnyi, S. G. Pavlostathis, and A. Rozzi, ‘Anaerobic Digestion Model No 1 ( ADM1 )’, Water Sci. Technol., vol. 45, no. 10, pp. 65–73, 2002.
Sh.-S. Jeong, C.-W. Suh, J.-L. Lim, S.-H. Lee, and H.-S. Shin, ‘Analysis and application of ADM1 for anaerobic methane production’, Bioprocess Biosyst. Engenieering, vol. 27, pp. 81–89, 2005, doi: 10.1007/s00449-004-0370-4.
D. G. Ibarlucía, E. M. Santalla, and V. E. Córdoba, ‘Evaluation of biomethane potential and kinetics modelling of green macroalgae from the South Atlantic Sea: Codium sp. (Codiaceae) and Ulva sp. (Ulvaceae)’, Environ. Chem., vol. 18, no. 7, pp. 311–320, 2021, doi: 10.1071/EN21088.
D. D. Nguyen et al., ‘Thermophilic anaerobic digestion of model organic wastes : Evaluation of biomethane production and multiple kinetic models analysis’, Bioresour. Technol., vol. 280, no. December 2018, pp. 269–276, 2019, doi: 10.1016/j.biortech.2019.02.033.
Z. Mou, C. Scheutz, and P. Kjeldsen, ‘Evaluating the methane generation rate constant (k value) of low-organic waste at Danish landfills’, Waste Manag., vol. 35, pp. 170–176, 2015, doi: 10.1016/j.wasman.2014.10.003.
R. S. Monzamodeth, J. F. Flores-Alvarez, I. Reyes-Chaparro, F. Castillo, B. Campillo, and O. Flores, ‘Artificial neural networks for predicting potentiodynamic tests of brass 70-30’, Mater. Today Proc., no. xxxx, pp. 1–5, 2023, doi: 10.1016/j.matpr.2023.01.287.
F. Ahmed and W. Chen, ‘Investigation of steam ejector parameters under three optimization algorithm using ANN’, Appl. Therm. Eng., vol. 225, no. December 2022, p. 120205, 2023, doi: 10.1016/j.applthermaleng.2023.120205.
T. Beltramo, C. Ranzan, J. Hinrichs, and B. Hitzmann, ‘Artificial neural network prediction of the biogas flow rate optimised with an ant colony algorithm’, Biosyst. Eng., vol. 143, pp. 68–78, 2016, doi: 10.1016/j.biosystemseng.2016.01.006.
F. Tufaner and Y. Demirci, ‘Prediction of biogas production rate from anaerobic hybrid reactor by artificial neural network and nonlinear regressions models’, Clean Technol. Environ. Policy, no. 0123456789, 2020, doi: 10.1007/s10098-020-01816-z.
R. Alrowais, A. Al-otaibi, A. Y. Hatata, M. A. Essa, and M. M. Abdel, ‘Comparing the effect of mesophilic and thermophilic anaerobic co-digestion for sustainable biogas production : An experimental and recurrent neural network model study’, J. Clean. Prod., vol. 392, no. December 2022, p. 136248, 2023, doi: 10.1016/j.jclepro.2023.136248.
D. Strik, A. Domnanovich, L. Zani, R. Braun, and P. Holubar, ‘Prediction of trace compounds in biogas from anaerobic digestion using the MATLAB Neural Network Toolbox’, Environ. Model. Softw., vol. 20, no. 6, pp. 803–810, Jun. 2005, doi: 10.1016/j.envsoft.2004.09.006.
M. Das Ghatak and A. Ghatak, ‘Artificial neural network model to predict behavior of biogas production curve from mixed lignocellulosic co-substrates’, Fuel, vol. 232, no. May 2017, pp. 178–189, 2018, doi: 10.1016/j.fuel.2018.05.051.
V. V Nair, H. Dhar, S. Kumar, A. Kumar, S. Mukherjee, and J. W. C. Wong, ‘Artificial neural network based modeling to evaluate methane yield from biogas in a laboratory-scale anaerobic bioreactor’, Bioresour. Technol., vol. 217, pp. 90–99, 2016, doi: 10.1016/j.biortech.2016.03.046.
APHA, APHA: Standard Methods for the Examination of Water and Wastewater. 20th ed. Washington, DC., no. 1. American Public Health Association, 1999.
S. Jenkins, J. Morgan, and C. Sawyer, ‘Measuring anaerobic sludge digestion and growth by a simple alkalimetric titration’, Water Pollut. Control Fed., vol. 55, no. 5, pp. 448–453, 1983, [Online]. Available: http://www.jstor.org/stable/25041903.
C. Lv et al., ‘Levenberg-marquardt backpropagation training of multilayer neural networks for state estimation of a safety-critical cyber-physical system’, IEEE Trans. Ind. Informatics, vol. 14, no. 8, pp. 3436–3446, 2018, doi: 10.1109/TII.2017.2777460.
I. Angelidaki, L. Ellegaard, and B. K. Ahring, ‘A Mathematical Model for Dynamic Simulation of Anaerobic Digestion of Complex Substrates: Focusing on Ammonia Inhibition’, Biotechnol. Bioeng., vol. 42, pp. 159–166, 1993, doi: 10.1002/bit.260420203.
F. Almomani, ‘Prediction of biogas production from chemically treated co-digested agricultural waste using arti fi cial neural network’, Fuel, vol. 280, no. July, p. 118573, 2020, doi: 10.1016/j.fuel.2020.118573.
A. Hublin and B. Zelic, ‘Modelling of the whey and cow manure co-digestion process.’, Waste Manag. Res. J. Int. Solid Wastes Public Clean. Assoc. ISWA, vol. 31, no. 4, pp. 353–60, Apr. 2013, doi: 10.1177/0734242X12455088.
F. M. Pellera and E. Gidarakos, ‘Effect of substrate to inoculum ratio and inoculum type on the biochemical methane potential of solid agroindustrial waste’, J. Environ. Chem. Eng., vol. 4, pp. 3217–3229, 2016, doi: 10.1016/j.jece.2016.05.026.
M. H. Gerardi, The microbiology of anaerobic digesters, 1st ed. John Wiley & Sons, Inc., Hoboken, New Jersey, 2003.
T. H. Ergüder, U. Tezel, E. Güven, and G. N. Demirer, ‘Anaerobic biotransformation and methane generation potential of cheese whey in batch and UASB reactors.’, Waste Manag., vol. 21, no. 7, pp. 643–50, Jan. 2001, [Online]. Available: http://www.ncbi.nlm.nih.gov/pubmed/11530920.
B. Lagoa-Costa, C. Kennes, and M. C. Veiga, ‘Cheese whey fermentation into volatile fatty acids in an anaerobic sequencing batch reactor’, Bioresour. Technol., vol. 308, no. March, p. 123226, 2020, doi: 10.1016/j.biortech.2020.123226.