Estimation of Reproduction Number for Covid-19 with Statistical Correction of Notifications Delay
Keywords:
Reproduction number, Deconvolution, Notifications delay, Covid-19, Sars-CoV-2Abstract
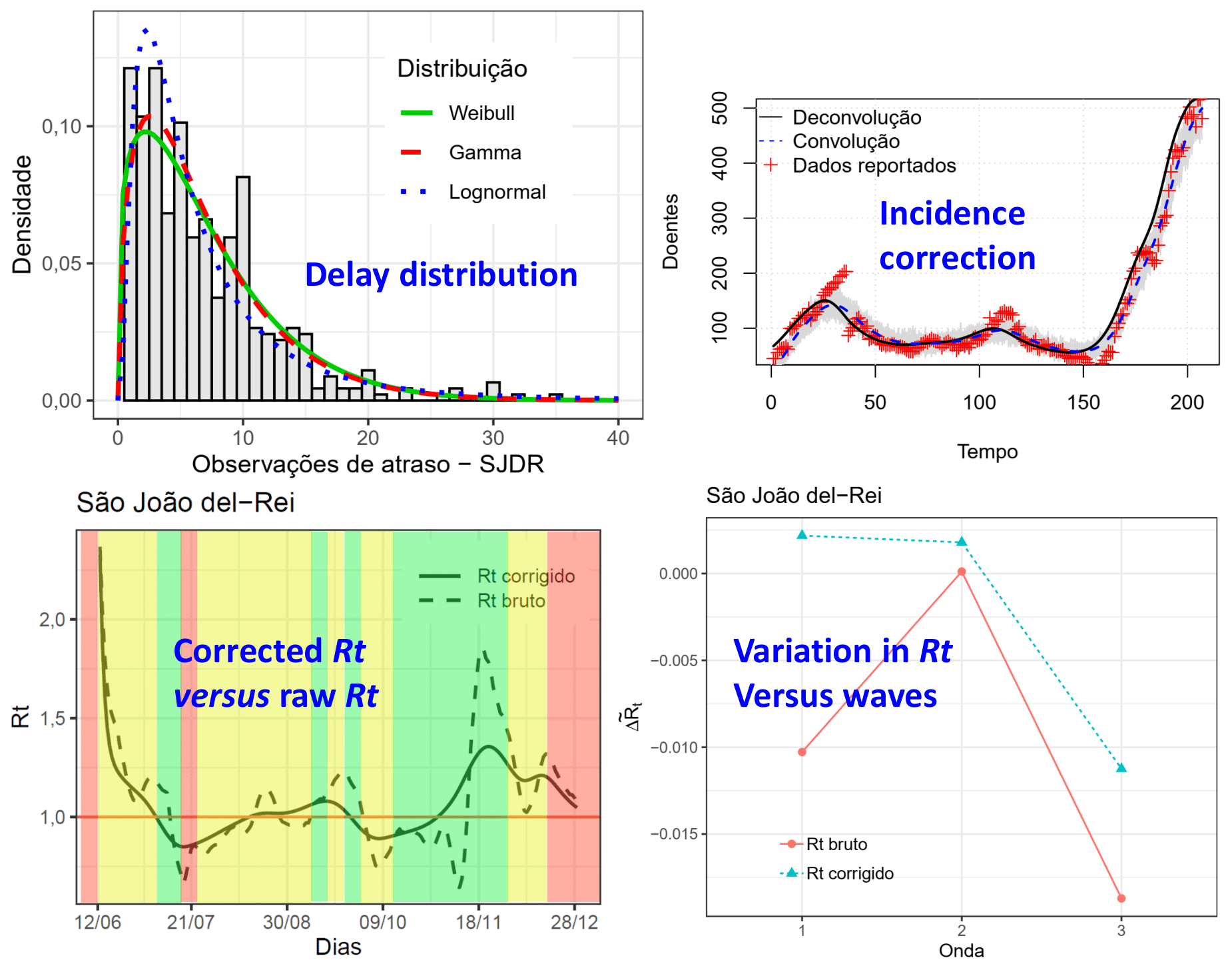
The pandemic of Covid-19 began in Brazil in February 2020. To evaluate the evolution of pandemics some metrics can be estimated, such as the reproduction number, Rt, and the basic reproduction number, R0. Due to the delay in the notifications, these estimates may present a bias. Taking the reported data, besides a sample of individuals who reported the day of symptoms onset, it is possible to estimate delay probabilities and to perform a deconvolution to correct the notifications' delay. In this work, it was performed a corrected estimate of Rt. This estimate is done based on the curve of notifications corrected through deconvolution. The approach is applied in three country cities and in the capital of Minas Gerais state. The behavior of Rt concerning the Minas Consciente program was evaluated. It was observed that the corrected Rt was more suitable to measure the effect of the program when compared to the raw Rt. When it was determined a more rigid mobility and activities regime by the program, it was observed a decrease in the median of the variation of the Rt of the cities studied.
Downloads
References
Ministério da Saúde, “Sobre a doença - coronavírus.” https://coronavirus.saude.gov.br/sobre-a-doenca. Acesso em: 02
mar. 2021.
D. Pasetto, J. C. Lemaitre, E. Bertuzzo, M. Gatto, and A. Rinaldo, “Range of reproduction number estimates for covid-19 spread.” 2020.
S. Wang, X. Yang, L. Li, P. Nadler, R. Arcucci, Y. Huang, Z. Teng, and Y. Guo, “A bayesian updating scheme for pandemics: Estimating
the infection dynamics of covid-19,” IEEE Computational Intelligence Magazine, vol. 15, no. 4, pp. 23–33, 2020.
I. McCulloh, K. Kiernan, and T. Kent, “Improved estimation of daily covid-19 rate from incomplete data,” in 2020 Fourth International
Conference on Multimedia Computing, Networking and Applications (MCNA), pp. 153–158, IEEE, 2020.
Governo de Minas Gerais, “Minas consciente - entenda o plano.” https://www.mg.gov.br/minasconsciente/entenda-o-programa. Acesso em: 11 maio 2021.
A. J. Sarnaglia, B. Zamprogno, F. A. F. Molinares, L. G. de Godoi, and N. A. J. Monroy, “Correcting notification delay and forecasting of covid-19 data,” Journal of mathematical analysis and applications, p. 125202, 2021.
D. A. M. Villela, “How limitations in data of health surveillance impact decision making in the covid-19 pandemic,” Saúde em debate, pp. 206–2018.
O. N. Bjornstad, Epidemics: Models and Data using R. Switzerland, AG: Springer International Publishing, 2018.
V. López and M. Cuki ˇ c, “A dynamical model of sars-cov-2 based on ´people flow networks,” Safety Science, vol. 134, p. 105034, 2021.
A. Cori, N. M. Ferguson, C. Fraser, and S. Cauchemez, “A New Framework and Software to Estimate Time-Varying Reproduction Numbers During Epidemics,” American Journal of Epidemiology, vol. 178, pp. 1505–1512, 09 2013.
H. Nishiura, N. M. Linton, and A. R. Akhmetzhanov, “Serial interval of novel coronavirus (covid-19) infections,” International Journal of
Infectious Diseases, vol. 93, pp. 284–286, 2020.
A. C. Miller, L. Hannah, J. Futoma, N. J. Foti, E. B. Fox, A. D’Amour, M. Sandler, R. A. Saurous, and J. Lewnard, “Statistical deconvolution
for inference of infection time series,” medRxiv, 2020.
E. Goldstein, J. Dushoff, J. Ma, J. B. Plotkin, D. J. Earn, and M. Lipsitch, “Reconstructing influenza incidence by deconvolution of daily mortality time series,” Proceedings of the National Academy of Sciences, vol. 106, no. 51, pp. 21825–21829, 2009.
S. Wang, X. Yang, L. Li, P. Nadler, R. Arcucci, Y. Huang, Z. Teng, and Y. Guo, “A bayesian updating scheme for pandemics: estimating
the infection dynamics of covid-19,” IEEE Computational Intelligence Magazine, vol. 15, no. 4, pp. 23–33, 2020.
Wesley Cota, PhD, “Número de casos confirmados de covid-19 no Brasil.” https://github.com/wcota/covid19br. Acesso em: 18 maio 2020.
Governo Federal, “Opendatasus.” https://opendatasus.saude.gov.br/dataset/bd-srag-2021. Acesso em: 05 mar. 2021.
Governo de Minas Gerais, “Minas consciente - transparência.” https://www.mg.gov.br/minasconsciente/transparencia. Acesso em: 19 maio 2021.
F. Najafi, N. Izadi, S.-S. Hashemi-Nazari, F. Khosravi-Shadmani, R. Nikbakht, and E. Shakiba, “Serial interval and time-varying reproduction number estimation for covid-19 in western iran,” New Microbe and New Infect, vol. 36, no. 100715, 2020.
Q. Li, X. Guan, P. Wu, X. Wang, L. Zhou, Y. Tong, R. Ren, K. S. Leung, E. H. Lau, J. Y. Wong, et al., “Early transmission dynamics in
wuhan, china, of novel coronavirus–infected pneumonia,” New England journal of medicine, 2020.
V. Viego, M. Geri, J. Castiglia, and E. Jouglard, “Período de incubación e intervalo serial para covid-19 en una cadena de transmisión en bahia blanca (argentina),” Ciência Saúde Coletiva, vol. 25, no. 9, 2020.
S. Zhao, D. Gao, Z. Zhuang, M. K. Chong, Y. Cai, J. Ran, P. Cao, K. Wang, Y. Lou, W. Wang, et al., “Estimating the serial interval of
the novel coronavirus disease (covid-19): A statistical analysis using the public data in hong kong from january 16 to february 15, 2020,” 2020.
M. L. Delignette-Muller and C. Dutang, “fitdistrplus: An R package for fitting distributions,” Journal of Statistical Software, vol. 64, no. 4, pp. 1–34, 2015.
A. Cori, EpiEstim: Estimate Time Varying Reproduction Numbers from Epidemic Curves, 2019. R package version 2.2-1.
A. Miller, L. Hannah, N. Foti, and J. Futoma, incidental: Implements Empirical Bayes Incidence Curves, 2020. R package version 0.1.
W. M. de Souza, L. F. Buss, D. da Silva Candido, J.-P. Carrera, S. Li, A. E. Zarebski, R. H. M. Pereira, C. A. Prete, A. A. de Souza-Santos,
K. V. Parag, et al., “Epidemiological and clinical characteristics of the covid-19 epidemic in brazil,” Nature Human Behaviour, vol. 4, no. 8,
pp. 856–865, 2020.
E. Shim, A. Tariq, and G. Chowell, “Spatial variability in reproduction number and doubling time across two waves of the covid-19 pandemic in south korea, february to july, 2020,” International Journal of Infectious Diseases, vol. 102, pp. 1–9, 2021.