An End-to-End Deep Learning System for Hop Classification
Keywords:
Hop, Convolutional neural network, Leaf recognition, Data augmentationAbstract
Automatic classification of plant species is a very challenging and widely studied problem in the literature. Distinguishing different varieties within the same species is an even more challenging task although less explored. Nevertheless, for some species distinguishing the varieties within the species can be of paramount importance.
Hops, a plant widely used in beer production, has over 250 cataloged varieties. Although the varieties have similar appearances, their chemical components, which influence the aroma and flavor of the drink, are quite heterogeneous. Therefore, it is important for producers to distinguish which variety the plant belongs to in a simple manner.
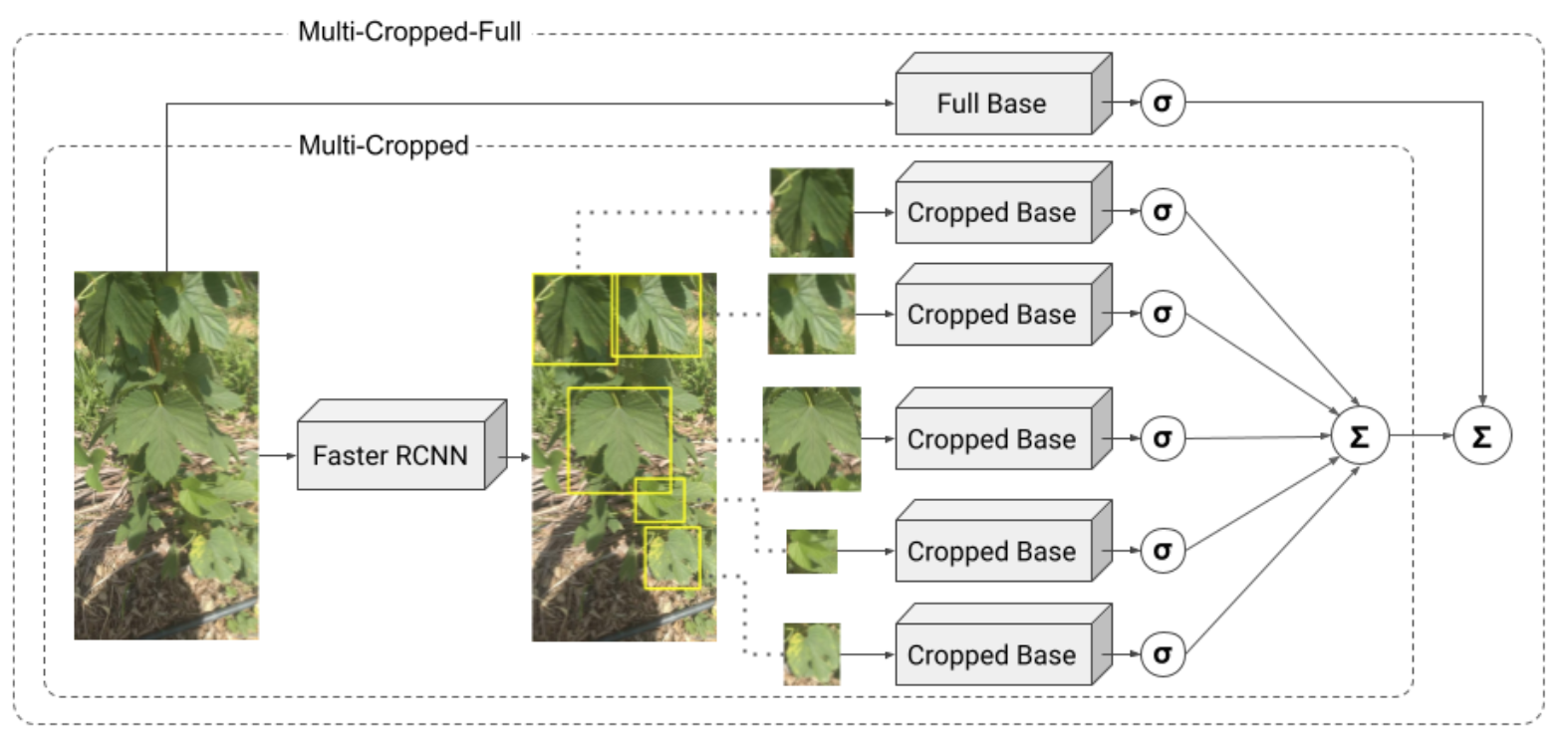
In this work, an end-to-end deep learning system is proposed to automate the task of hop classification. Five architectures are proposed and evaluated with an uncontrolled environment dataset that includes 12 varieties of hops on 1592 images, from three different cell phone cameras. The best architecture automatically detects the hop leaves on the image and performs the classification using the information of up to 10 leaves. The method achieved an accuracy of 95.69% with an inference time of 672ms. To reach such figures, state-of-the-art convolutional blocks were explored along with data augmentation techniques. Our results show that the system is robust and has a low computational cost.
Downloads
References
G. Astray, P. Gullón, B. Gullón, P. E. Munekata, and J. M. Lorenzo, “Humulus lupulus l. as a natural source of functional biomolecules,” Applied Sciences , vol. 10, no. 15, p. 5074, 2020.
C. Almaguer, C. Schönberger, M. Gastl, E. K. Arendt, and T. Becker, “Humulus lupulus–a story that begs to be told. a review,”
Journal of the Institute of Brewing , vol. 120, no. 4, pp. 289–314, 2014.
B. Kramer, J. Thielmann, A. Hickisch, P. Muranyi, J. Wunderlich, and C. Hauser, “Antimicrobial activity of hop extracts against foodborne pathogens for meat applications,” Journal of applied microbiology ,vol. 118, no. 3, pp. 648–657, 2015.
T. Nuutinen, “Medicinal properties of terpenes found in cannabis sativa and humulus lupulus,” European journal of medicinal chemistry, vol. 157, pp. 198–228, 2018.
L. Chadwick, G. Pauli, and N. Farnsworth, “The pharmacognosy of humulus lupulus l.(hops) with an emphasis on estrogenic properties,” Phytomedicine , vol. 13, no. 1-2, pp. 119–131, 2006.
P. Zanoli and M. Zavatti, “Pharmacognostic and pharmacological profile of humulus lupulus l.,” Journal of ethnopharmacology
, vol. 116, no. 3, pp. 383–396, 2008.
M. Biendl and C. Pinzl, “Hops and health,” MBAA TQ , vol. 46, pp. 1–7, 2009.
M. Van Cleemput, K. Cattoor, K. De Bosscher, G. Haegeman, D. De Keukeleire, and A. Heyerick, “Hop (humulus lupulus)-derived bitter acids as multipotent bioactive compounds,” Journal of natural products , vol. 72, no. 6, pp. 1220–1230, 2009.
J. Healey, The Hops List: 265 Beer Hop Varieties From Around the World. Julian Healey, 2016.
B. Steenackers, L. De Cooman, and D. De Vos, “Chemical transformations of characteristic hop secondary metabolites in relation to beer properties and the brewing process: a review,” Food Chemistry , vol. 172, pp. 742–756, 2015.
A. da Rosa Almeida, M. V. d. O. B. Maciel, B. Cardoso Gasparini Gandolpho, M. H. Machado, G. L. Teixeira, F. C. Bertoldi, C. M. Noronha, L. Vitali, J. M. Block, and P. L. M. Barreto, “Brazilian grown cascade hop (humulus lupulus l.): Lc-esi-ms-ms and gc-ms analysis of chemical composition and antioxidant activity of extracts and essential oils,” Journal of the American Society of Brewing Chemists , pp. 1–11, 2020.
A. d. R. Almeida, M. V. d. O. B. Maciel, M. H. Machado, G. C. Bazzo, R. D. de Armas, V. B. Vitorino, L. Vitali, J. M. Block, and P. L. M. Barreto, “Bioactive compounds and antioxidant activities of brazilian hop (humulus lupulus l.) extracts,” International Journal of Food Science & Technology , vol. 55, no. 1, pp. 340–347, 2020.
T. R. Arruda, P. F. Pinheiro, P. I. Silva, and P. C. Bernardes, “A new perspective of a well-recognized raw material: Phenolic content, antioxidant and antimicrobial activities and α-and β-acids profile of brazilian hop (humulus lupulus l.) extracts,” LWT , vol. 141, p. 110905,
L. De Cooman, E. Everaert, and D. De Keukeleire, “Quantitative analysis of hop acids, essential oils and flavonoids as a clue to the identification of hop varieties,” Phytochemical Analysis: An International Journal of Plant Chemical and Biochemical Techniques
, vol. 9, no. 3, pp. 145–150, 1998.
M. Kovacevic and M. Kac, “Determination and verification of hop varieties by analysis of essential oils,” Food Chemistry
, vol. 77, no. 4, pp. 489–494, 2002.
R. A. Shellie, S. D. Poynter, J. Li, J. L. Gathercole, S. P. Whittock,and A. Koutoulis, “Varietal characterization of hop (humulus lupulus l.)by gc–ms analysis of hop cone extracts,”Journal of separation science,vol. 32, no. 21, pp. 3720–3725, 2009.
M. A. Farag, E. A. Mahrous, T. Lübken, A. Porzel, and L. Wessjohann,“Classification of commercial cultivars of humulus lupulus l.(hop)by chemometric pixel analysis of two dimensional nuclear magnetic resonance spectra,”Metabolomics, vol. 10, no. 1, pp. 21–32, 2014.
L. M. Duarte, L. H. C. Adriano, and M. A. L. de Oliveira, “Capillaryelectrophoresis in association with chemometrics approach for bitterness hop (humulus lupulus l.) classification,”Electrophoresis, vol. 39, no. 11,pp. 1399–1409, 2018.
A. Kamilaris and F. X. Prenafeta-Boldú, “Deep learning in agriculture: A Survey,”Computers and electronics in agriculture, vol. 147, pp. 70–90,2018.
M. A. Jenks, “Plant nomenclature,”Purdue University-Department ofHorticulture and Landscape Architecture. Disponível, 2011.
N. Sünderhauf, C. McCool, B. Upcroft, and T. Perez, “Fine-grained plant classification using convolutional neural networks for feature extraction.,” in CLEF (Working Notes), pp. 756–762, 2014.
S. H. Lee, C. S. Chan, P. Wilkin, and P. Remagnino, “Deep-plant:Plant identification with convolutional neural networks,” in2015 IEEEinternational conference on image processing (ICIP), pp. 452–456,IEEE, 2015.
B. P. Tóth, M. J. Tóth, D. Papp, and G. Szücs, “Deep learning and svm classification for plant recognition in content-based large scale image retrieval.,” inCLEF (Working Notes), pp. 569–578, 2016.[24] H. Yalcin and S. Razavi, “Plant classification using convolutional neural networks,” in2016 Fifth International Conference on Agro-Geoinformatics (Agro-Geoinformatics), pp. 1–5, IEEE, 2016.
S. H. Lee, Y. L. Chang, C. S. Chan, and P. Remagnino, “Plant Identification system based on a convolutional neural network for the life clef 2016 plant classification task.,”CLEF (Working Notes), vol. 1,pp. 502–510, 2016.
Y. Sun, Y. Liu, G. Wang, and H. Zhang, “Deep learning for plant identification in natural environment,”Computational intelligence and neuroscience, vol. 2017, 2017.
W.-S. Jeon and S.-Y. Rhee, “Plant leaf recognition using a convolutional neural network,”International Journal of Fuzzy Logic and IntelligentSystems, vol. 17, no. 1, pp. 26–34, 2017.
J. Hu, Z. Chen, M. Yang, R. Zhang, and Y. Cui, “A multiscale fusion convolutional neural network for plant leaf recognition,”IEEE SignalProcessing Letters, vol. 25, no. 6, pp. 853–857, 2018.
X. Zhu, M. Zhu, and H. Ren, “Method of plant leaf recognition based on improved deep convolutional neural network,”Cognitive SystemsResearch, vol. 52, pp. 223–233, 2018.
B. Wang and D. Wang, “Plant leaves classification: A few-shot learning method based on siamese network,”IEEE Access, vol. 7, pp. 151754–151763, 2019.
Y. Zhu, W. Sun, X. Cao, C. Wang, D. Wu, Y. Yang, and N. Ye, “Ta-cnn: Two-way attention models in deep convolutional neural network for plant recognition,”Neurocomputing, vol. 365, pp. 191–200, 2019.
D. I. Mercurio and A. A. Hernandez, “Classification of sweet potato variety using convolutional neural network,” in2019 IEEE 9th International Conference on System Engineering and Technology (ICSET),pp. 120–125, IEEE, 2019.
K. Yang, W. Zhong, and F. Li, “Leaf segmentation and classification with a complicated background using deep learning,”Agronomy, vol. 10,no. 11, p. 1721, 2020.
M. Lin, Q. Chen, and S. Yan, “Network in network,”arXiv preprintarXiv:1312.4400, 2013.[35] C. Szegedy, V. Vanhoucke, S. Ioffe, J. Shlens, and Z. Wojna, “Rethinking The inception architecture for computer vision,” inProceedings of the IEEE conference on computer vision and pattern recognition, pp. 2818–2826, 2016.
K. He, X. Zhang, S. Ren, and J. Sun, “Deep residual learning for image recognition,” in Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 770–778, 2016.
M. Tan and Q. Le, “Efficientnet: Rethinking model scaling for con-volutional neural networks,” inInternational Conference on MachineLearning, pp. 6105–6114, PMLR, 2019.
S. Ren, K. He, R. Girshick, and J. Sun, “Faster r-cnn: towards real-time object detection with region proposal networks,”IEEE transactions on pattern analysis and machine intelligence, vol. 39, no. 6, pp. 1137–1149,2016.
T. DeVries and G. W. Taylor, “Improved regularization of convolutional neural networks with cutout,”arXiv preprint arXiv:1708.04552, 2017.
J. Duchi, E. Hazan, and Y. Singer, “Adaptive subgradient methods for online learning and stochastic optimization.,” Journal of machine learning research , vol. 12, no. 7, 2011.
I. Kandel, M. Castelli, and A. Popovic, “Comparative study of first order optimizers for image classification using convolutional neural networks on histopathology images,” Journal of Imaging , vol. 6, no. 9, p. 92, 2020.
X. Glorot and Y. Bengio, “Understanding the difficulty of training deep feedforward neural networks,” in Proceedings of the thirteenth international conference on artificial intelligence and statistics , pp. 249– 256, JMLR Workshop and Conference Proceedings, 2010.