G-CoV.2: A computational system for geolocation of patients diagnosed with COVID-19
Keywords:
Coronaviruses, Geographic information systems, Pandemics, PathologyAbstract
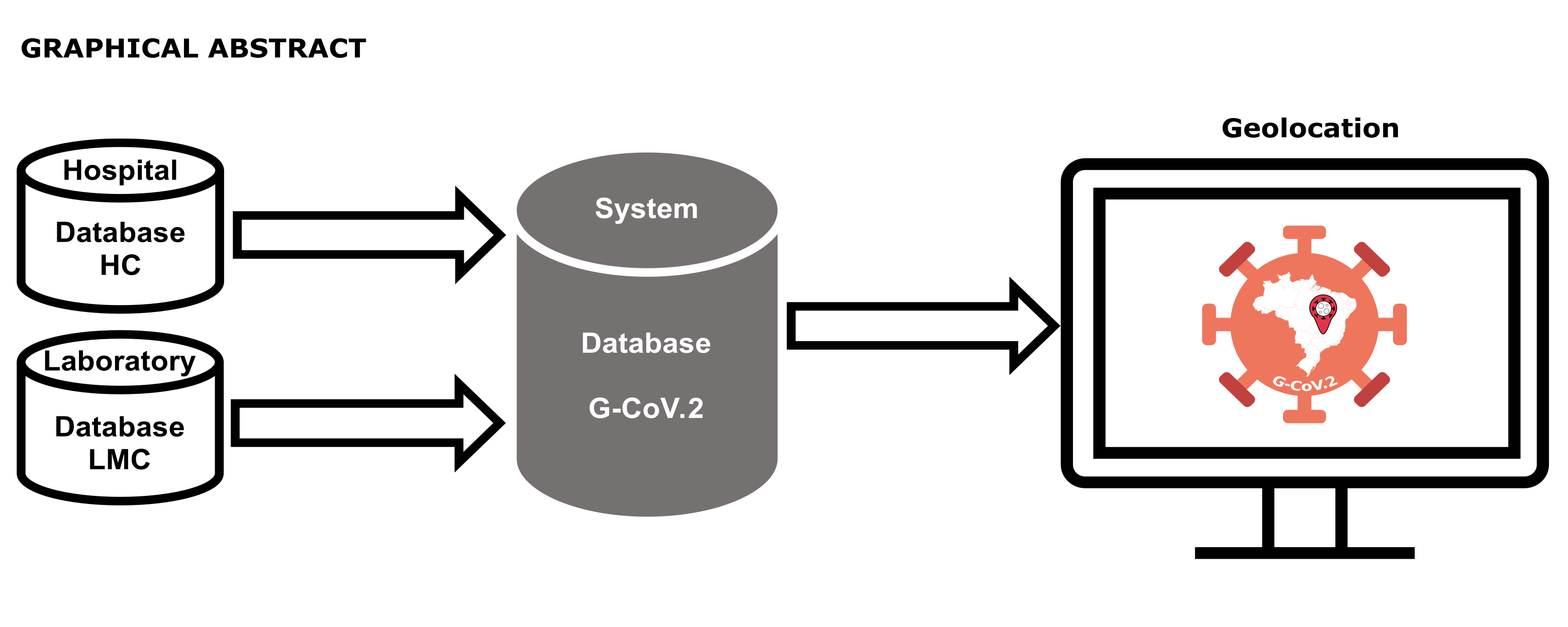
Coronavirus disease 2019 (COVID-19) is a respiratory infection caused by the Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV-2). In the current scenario of COVID-19, more than 6 million people have died worldwide, and near seven hundred thousand only in Brazil alone. The objective this work is designer a database using MySQL to develop a system for geolocating COVID-19 affected individuals and their close contacts. Patient addresses registered in the laboratory database were used to search for coordinate (latitude and longitude) using the OpenStreetMap Application Programming Interface. The user interface was developed in PHP using HTML, JavaScript, CSS, and AJAX. A dashboard displaying the geolocation information of selected patients was presented on the screen for public health agents. A system called G-CoV.2 was developed. The makers produced by G-CoV.2 had 100% accuracy when manually compared with the same patient address records. G-CoV.2 can be used as a computational system to aid health managers in identifying regions with the most people infected by COVID-19 or other pathological processes. The submitted work can meet the third goal of sustainable development of the United Nations for Latin America, ensuring a healthy life and thus promoting well-being for all people, at all ages. One of the ways is through the monitoring of COVID-19 and other pathologies as a preventive measure, applying the system developed G-CoV.2 that was the subject of the writing of the article, which may reduce the mortality rate at all ages, with the premise of ending epidemics of serious and communicable diseases, directing health managers to increase investment in health in critical areas, as well as creating actions in favor of prevention and promotion of the population's health.
Downloads
References
S. Ludwig and A. Zarbock, "Coronaviruses and SARS-CoV-2: A Brief Overview," Anesthesia & Analgesia, vol. 131, no. 1, pp. 93-96, Jul. 2020, doi: 10.1213/ANE.0000000000004845.
C. Huang et al., "Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China," The Lancet, vol. 395, no. 10223, pp. 497-506, 2020, doi: 10.1016/S0140-6736(20)30183-5.
Y.-F. Tu et al., "A Review of SARS-CoV-2 and the Ongoing Clinical Trials," International Journal of Molecular Sciences, vol. 21, no. 7, pp. 2657-2657, Apr. 2020, doi: 10.3390/ijms21072657.
D. Wang et al., "Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China," JAMA, vol. 323, no. 11, pp. 1061-1069, Mar. 2020, doi: 10.1001/jama.2020.1585.
J. Alizargar, "Risk of reactivation or reinfection of novel coronavirus (COVID-19)," Journal of the Formosan Medical Association, vol. 119, no. 6, pp. 1123-1123, Jun. 2020, doi: 10.1016/j.jfma.2020.04.013.
World Health Organization, "Critical preparedness, readiness and response actions for COVID-19: interim guidance, 22 March 2020," Geneva – SZ, 2020. Available from:
https://apps.who.int/iris/handle/10665/331511.
Ministério da Saúde, "Apêndice das Diretrizes para Diagnóstico e Tratamento da COVID-19," Brasília - DF, 2020. Avalaible from: https://rebrats.saude.gov.br/images/Documentos/Apendice_COVID.pdf
D. A. P. MIRANDA et al. "Long COVID-19 syndrome: a 14-months longitudinal study during the two first epidemic peaks in Southeast Brazil," Transactions of The Royal Society of Tropical Medicine and Hygiene, vol. 116, no. 11, pp. 1007-1014, May. 2022, doi: 10.1093/trstmh/trac030.
A. V. Raveendran, R. Jayadevan, and S. Sashidharan, "Long COVID: An overview," Diabetes & Metabolic Syndrome: Clinical Research & Reviews, vol. 15, no. 3, pp. 869-875, May. 2021, doi: 10.1016/j.dsx.2021.04.007.
Conselho das Organizações Internacionais de Ciências Médicas Médicas. "Diretrizes Éticas Internacionais para Pesquisas Relacionadas a Saúde Envolvendo Seres Humanos," Brasília - DF, 2018. Avalaible from: https://cioms.ch/wp-content/uploads/2018/11/CIOMS-final-Diretrizes-Eticas-Internacionais-Out18.pdf.
Ministério da Economia. "Brasil / Paraná / Curitiba: Panorama – Economia e Saúde." Rio de Janeiro – RJ, 2023. Available from: https://cidades.ibge.gov.br/brasil/pr/curitiba/panorama.
Ministério Da Educação. "Hospitais Universitários: Hospital de Clínicas," Curitiba – PR, 2020 Available from: https://www.gov.br/ebserh/pt-br/hospitais-universitarios/regiao-sul/chc-ufpr/acesso-a-informacao/institucional/hospital-de-clinicas
L. Signorini, "Extração e análise de dados de hemoglobina (HbA1c) como ferramenta de tomada de decisões estratégicas para o diabetes mellitus no Sistema Único de Saúde" Mestrado, Programa de Pós-Graduação em Ciências Farmacêuticas, Universidade Federal do Paraná, 2022.
Free Software Foundation, " The GNU Operating System and the Free Software Movement," Boston - MA, 2023. Available from: https://www.gnu.org/
F. Nelli, Python Data Analytics. Berkeley, CA: Apress Access Books, 2018, doi: 10.1007/978-1-4842-3913-1
P. P.-S. Chen, "The entity-relationship model—toward a unified view of data," ACM Transactions on Database Systems, vol. 1, no. 1, pp. 9-36, Mar. 1976, doi: 10.1145/320434.320440.
OpenStreetMap Foundation, "OpenStreetMap provides map data for thousands of websites, mobile apps, and hardware devices." Cambridge – UK, 2023. Available from: https://www.openstreetmap.org/about.
N. H. Rosa, P. U. Simioni, and B. Damião, "Evolução do SARS-CoV 2: comparação biológica da epidemia de SARS-CoV 1 com a pandemia de SARS-CoV 2," Revista da Faculdade de Ciências Médicas de Sorocaba, vol. 22, no. 4, pp. 141-145, Jun. 2022, doi: 10.23925/1984-4840.2020v22i4a2.
D. E. Services, HTML 5 Black Book, Covers CSS 3, JavaScript, XML, XHTML, AJAX, PHP and jQuery, 2nd ed. New Delhi: Dreamtech Press, 2016, p. 1252.
A. Prado et al., "G-CoV.2," BR512023000732-3 Mar. 21, 2023. pp. 20-21. http://revistas.inpi.gov.br/pdf/Programa_de_computador2724.pdf
P. Pérez Sust et al., "Turning the Crisis Into an Opportunity: Digital Health Strategies Deployed During the COVID-19 Outbreak," JMIR Public Health and Surveillance, vol. 6, no. 2, pp. e19106-e19106, May. 2020, doi: 10.2196/19106.
F. Almeida, "Technological Solutions for Screening and Geolocation of COVID-19 Infected People: A Country Comparison Approach," Frontiers in Health Informatics, vol. 10, no. 62, pp. 1-6, Feb. 2021, doi: 10.30699/fhi.v10i1.270.
Congresso Nacional. "Lei Geral de Proteção de Dados Pessoais (LGPD)," Brasília – DF, 2018. Available from: https://www.planalto.gov.br/ccivil_03/_ato2015-2018/2018/lei/l13709.htm.
J. González-Cabañas et al., "Digital Contact Tracing: Large-Scale Geolocation Data as an Alternative to Bluetooth-Based Apps Failure," Electronics, vol. 10, no. 9, May. 2021, doi: 10.3390/electronics10091093.
B. Mitrică et al., "Population Vulnerability to the SARS‐CoV‐2 Virus Infection. A County‐Level Geographical‐Methodological Approach in Romania," GeoHealth, vol. 5, no. 11, Nov. 2021, doi: 10.1029/2021GH000461.
M. C. Febriansyah et al., "An Android-based COVID Case Monitoring Application Using a Geographic Information System to Educate the Public to be More Aware of COVID-19 Spreads,” IOP Publishing, vol. 1158, pp. 012009, Jun. 2021, doi:10.1088/1757-899X/1158/1/012009.
A. M. K. Cheng, "Real-Time COVID-19 Infection Risk Assessment and Mitigation based on Public-Domain Data," 2021 IEEE/ACM HPC for Urgent Decision Making (UrgentHPC), pp. 29-35, Nov. 2021, doi: 10.1109/UrgentHPC54802.2021.00009.
P. G. Malpani, K. M. Shinde and M. R. Nalavade, "Location Proximity of Covid-19 Suspect Using Mobile Data History," 2020 IEEE Bangalore Humanitarian Technology Conference (B-HTC), pp. 1-5, Oct. 2020, doi: 10.1109/B-HTC50970.2020.9297838.
M. T. Rahman et al., "An Automated Contact Tracing Approach for Controlling Covid-19 Spread Based on Geolocation Data From Mobile Cellular Networks," IEEE Access, vol. 8, pp. 213554-213565, Nov. 2020, doi: 10.1109/ACCESS.2020.3040198.
R. Mahanthesh et al., "COVID-19 Contact Tracing Using Geolocation," Springer Singapore, vol. 2, no. 790, pp. 967-980. Dec. 2021, doi: 10.1007/978-981-16-1342-5_77.
M. Shahroz et al., "COVID-19 digital contact tracing applications and techniques: A review post initial deployments," Transportation Engineering, vol. 5, p. 100072, Sep. 2021, doi: https://doi.org/10.1016/j.treng.2021.100072.
R. Md. Tanvir, S. Taslima Ferdaus, K. Risala Tasin, and N. Mostofa Kamal, "A Review of Contact Tracing Approaches for Controlling COVID-19 Pandemic," Global Journal of Computer Science and Technology, vol. 21, no. H1, pp. 31-38, Jan. 2021, doi: 10.34257/GJCSTHVOL21IS1PG31.
N. Min-Allah et al., "A survey of COVID-19 contact-tracing apps," Computers in Biology and Medicine, vol. 137, p. 104787, Oct. 2021, doi: https://doi.org/10.1016/j.compbiomed.2021.104787.
H. Kondylakis et al., "COVID-19 Mobile Apps: A Systematic Review of the Literature," J Med Internet Res, vol. 22, no. 12, p. e23170, Dec. 2020, doi: 10.2196/23170.
N. Beeching and T. Fletcher, Fowler, R. "Coronavirus Disease 2019 (COVID-19) - Symptoms, diagnosis, and treatment." BMJ Best Practice. London - UK, 2023. Available from: https://bestpractice.bmj.com/topics/en-gb/3000201
Z. Wu and J. M. McGoogan, "Characteristics of and Important Lessons From the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72 314 Cases From the Chinese Center for Disease Control and Prevention," JAMA, vol. 323, no. 13, pp. 1239-1242, Apr. 2020, doi: 10.1001/jama.2020.2648.
W. Puryear et al., "Highly Pathogenic Avian Influenza A(H5N1) Virus Outbreak in New England Seals, United States," Emerg Infect Dis, vol. 29, no. 4, pp. 786-791, Apr. 2023, doi: 10.3201/eid2904.221538.
Centers for Disease Control and Prevention, "Long COVID or Post-COVID Conditions," Atlanta - GA, 2023. Available from: https://www.cdc.gov/coronavirus/2019-ncov/long-term-effects/index.html
S. J. Russell and P. Norvig, Artificial Intelligence: A Modern Approach, 4th ed. New Jersey: Pearson Education, Inc., 2021, p. 1168.