GeneConnector: Unlocking the full potential of Genbank metadata
Keywords:
Genbank, NCBI, Mycology, Phytopathology, GOPHY, GenomicsAbstract
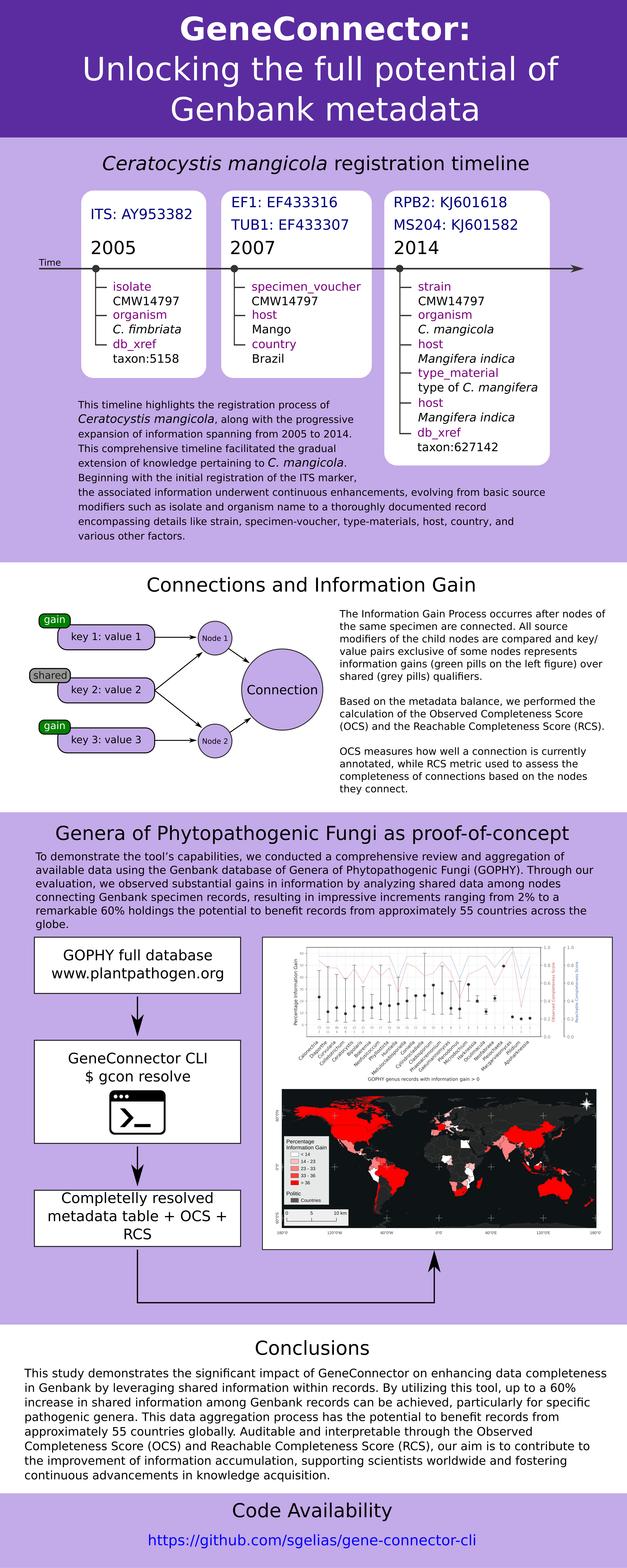
Genbank currently stands as one of the most significant global repositories of genetic information. However, despite its vast quantity and diversity of data, a considerable portion of the existing records suffer from disjointed and often lacking metadata, failing to provide the necessary context of their acquisition. In light of this, we propose GeneConnector, a tool that harnesses shared information among multiple records of the same specimen in Genbank, aiming to enhance the completeness of poorly annotated nodes across various information domains. To demonstrate the tool’s capabilities, we conducted a comprehensive review and aggregation of available data using the Genbank database of Genera of Phytopathogenic Fungi (GOPHY). Through our evaluation, we observed substantial gains in information by analyzing shared data among nodes connecting Genbank specimen records, resulting in impressive increments ranging from 2% to a remarkable 60%. Our approach empowers users to make precise, straightforward, and accurate assessments of the context associated to results, facilitated by two metrics that gauge the current level of data annotation and the potential information gain achievable following our evaluation.
Downloads
References
J. Shendure, G. M. Findlay, and M. W. Snyder, “Genomic medicine–progress, pitfalls, and promise,” Cell, vol. 177, no. 1, pp. 45–57, 2019.
R. Jeyasri, P. Muthuramalingam, L. Satish, S. K. Pandian, J.-T. Chen, S. Ahmar, X. Wang, F. Mora-Poblete, and M. Ramesh, “An overview of abiotic stress in cereal crops: negative impacts, regulation, biotechnology and integrated omics,” Plants, vol. 10, no. 7, p. 1472, 2021.
C. Juma, The gene hunters: Biotechnology and the scramble for seeds, vol. 996. Princeton University Press, 2014.
E. J. Gilchrist, S. Wang, and T. D. Quilichini, “The impact of biotechnology and genomics on an ancient crop: Cannabis sativa,” in Genomics and the Global Bioeconomy, pp. 177–204, Elsevier, 2023.
D. A. Benson, M. Cavanaugh, K. Clark, I. Karsch-Mizrachi, D. J. Lipman, J. Ostell, and E. W. Sayers, “Genbank,” Nucleic acids research, vol. 41, no. D1, pp. D36–D42, 2012.
K. L. Howe, P. Achuthan, J. Allen, J. Allen, J. Alvarez-Jarreta, M. R. Amode, I. M. Armean, A. G. Azov, R. Bennett, J. Bhai, et al., “Ensembl 2021,” Nucleic acids research, vol. 49, no. D1, pp. D884–D891, 2021.
U. Consortium, “Uniprot: a worldwide hub of protein knowledge,”Nucleic acids research, vol. 47, no. D1, pp. D506–D515, 2019.
D. Smedley, S. Haider, B. Ballester, R. Holland, D. London, G. Thorisson, and A. Kasprzyk, “Biomart–biological queries made easy,” BMC genomics, vol. 10, no. 1, pp. 1–12, 2009.
M. Van Wyk, B. D. Wingfield, A. O. Al-Adawi, C. J. Rossetto, M. F. Ito, and M. J. Wingfield, “Two new ceratocystis species associated with mango disease in brazil,” Mycotaxon, vol. 117, no. 1, pp. 381–404, 2011.
M. Van Wyk, A. Al-Adawi, B. Wingfield, A. Al-Subhi, M. Deadman, and M. Wingfield, “Dna based characterization of ceratocystis fimbriata isolates associated with mango decline in oman,” Australasian Plant Pathology, vol. 34, pp. 587–590, 2005.
M. Van Wyk, A. O. Al Adawi, I. A. Khan, M. L. Deadman, A. A. Al Jahwari, B. D. Wingfield, R. Ploetz, and M. J. Wingfield, “Ceratocystis manginecans sp. nov., causal agent of a destructive mango wilt disease in oman and pakistan,” Fungal Divers, vol. 27, pp. 213–230, 2007.
A. Fourie, M. J. Wingfield, B. D. Wingfield, and I. Barnes, “Molecular markers delimit cryptic species in ceratocystis sensu stricto,” Mycological Progress, vol. 14, pp. 1–18, 2015.
A. Canakoglu, A. Bernasconi, A. Colombo, M. Masseroli, and S. Ceri, “Genosurf: metadata driven semantic search system for integrated genomic datasets,” Database, vol. 2019, 2019.
Z. Chen, A. S. Azman, X. Chen, J. Zou, Y. Tian, R. Sun, X. Xu, Y. Wu, W. Lu, S. Ge, et al., “Global landscape of sars-cov-2 genomic surveillance and data sharing,” Nature genetics, vol. 54, no. 4, pp. 499–507, 2022.
U. Kõljalg, K.-H. Larsson, K. Abarenkov, R. H. Nilsson, I. J. Alexander, U. Eberhardt, S. Erland, K. Høiland, R. Kjøller, E. Larsson, et al., “Unite: a database providing web-based methods for the molecular identification of ectomycorrhizal fungi,” New Phytologist, vol. 166, no. 3, pp. 1063–1068, 2005.
K. Abarenkov, R. H. Nilsson, K.-H. Larsson, I. J. Alexander, U. Eberhardt, S. Erland, K. Høiland, R. Kjøller, E. Larsson, T. Pennanen, et al., “The unite database for molecular identification of fungi–recent updates and future perspectives,” The New Phytologist, vol. 186, no. 2, pp. 281–285, 2010.
M. Quiñones, D. T. Liou, C. Shyu, W. Kim, I. Vujkovic-Cvijin, Y. Belkaid, and D. E. Hurt, “Metagenote: a simplified web platform for metadata annotation of genomic samples and streamlined submission to ncbi’s sequence read archive,” BMC bioinformatics, vol. 21, pp. 1–12, 2020.
Á. Gálvez-Merchán, K. H. Min, L. Pachter, and A. S. Booeshaghi, “Metadata retrieval from sequence databases with ffq,” Bioinformatics, vol. 39, no. 1, p. btac667, 2023.
Q. Chen, J. Zobel, and K. Verspoor, “Duplicates, redundancies and inconsistencies in the primary nucleotide databases: a descriptive study,” Database, vol. 2017, 2017.
S. Reining, F. Ahlemann, B. Mueller, and R. Thakurta, “Knowledge accumulation in design science research: ways to foster scientific progress,” ACM SIGMIS Database: the DATABASE for Advances in Information Systems, vol. 53, no. 1, pp. 10–24, 2022.
Y. Marin-Felix, J. Groenewald, L. Cai, Q. Chen, S. Marincowitz, I. Barnes, K. Bensch, U. Braun, E. Camporesi, U. Damm, et al., “Genera of phytopathogenic fungi: Gophy 1,” Studies in mycology, vol. 86, pp. 99–216, 2017.
Y. Marin-Felix, M. Hernández-Restrepo, M. J. Wingfield, A. Akulov, A. Carnegie, R. Cheewangkoon, D. Gramaje, J. Z. Groenewald, V. Guarnaccia, F. Halleen, et al., “Genera of phytopathogenic fungi: Gophy 2,” Studies in mycology, vol. 92, pp. 47–133, 2019.
Y. Marin-Felix, M. Hernández-Restrepo, I. Iturrieta-González, D. García, J. Gené, J. Z. Groenewald, L. Cai, Q. Chen, W. Quaedvlieg, R. Schumacher, et al., “Genera of phytopathogenic fungi: Gophy 3,” Studies in mycology, vol. 94, pp. 1–124, 2019.
Q. Chen, M. Bakhshi, Y. Balci, K. Broders, R. Cheewangkoon, S. Chen, X. Fan, D. Gramaje, F. Halleen, M. Horta Jung, et al., “Genera of phytopathogenic fungi: Gophy 4,” Studies in Mycology, vol. 101, no. 1, pp. 417–564, 2022.
G. Van Rossum and F. L. Drake, Python 3 Reference Manual. Scotts Valley, CA: CreateSpace, 2009.
A. Cockburn, “Ports and adapters architecture,” 2006. http://wiki.c2.com/?PortsAndAdaptersArchitecture [Accessed: 2022-11-20].
D. Merkel, “Docker: lightweight linux containers for consistent development and deployment,” Linux journal, vol. 2014, no. 239, p. 2, 2014.