Review of Intelligent Algorithms for Breast Cancer Control: a Latin America Perspective
Keywords:
Artificial intelligence, Machine learning, Breast cancer, Systematic review, Meta-analysisAbstract
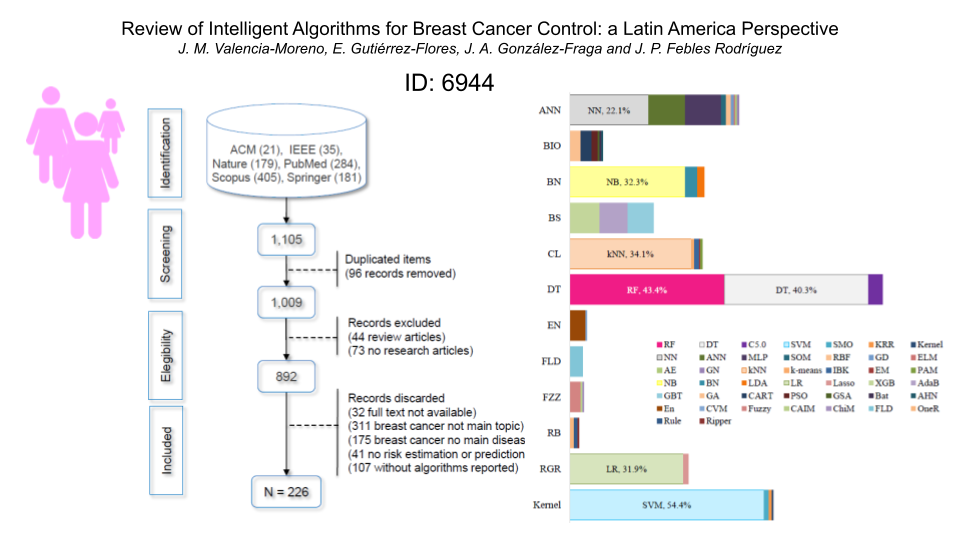
Breast cancer in women is a worldwide health problem that is one of the main causes of death. This situation is accentuated in Latin America and the Caribbean countries, where about 159 women die daily from this disease. The World Health Organization recommends focusing on Prevention and Early Detection of cancer to reduce mortality. However, this requires a great deal of information processing and analysis by experts, who require the support of technology to perform these tasks promptly. In recent years, the use of so-called intelligent algorithms has increased to support the fight against breast cancer. The authors summarized the studies published between January 2016 and June 2021, highlighting the current situation and opportunities for Latin America and the Caribbean. Studies were selected using the following terms: intelligent algorithms, assessment metrics, stages of breast cancer control addressed, data sources, data types, female population with breast cancer under study and the countries of the authors who have written articles on this subject. In this study, after applying the inclusion and exclusion criteria 226 articles were selected from a total of 1,105 articles found in the ACM digital library, IEEE Explore, Nature, PubMed, Scopus (Science Direct) and Springer Link databases. Publication between January 2016 and June 2021, breast cancer as main interest, algorithm and data type information, along with compliance with the general question were the inclusion criteria, while, being a research article, compliance with the three subqueries and availability, were the exclusion criteria. Using a spreadsheet as based tool to collect and analyze the data, the study found that the most used elements were: SVM, RF and DT algorithms; accuracy as assessment metric; public information sources; data on tumors (size and shape, among others); USA information sources; India as the country of the first authors who wrote the most articles of the selected papers; and Diagnosis & Treatment as the most addressed stage of cancer control. Results in this review paper provide an overview of the application of intelligent algorithms against breast cancer. In this regard, the gaps that were detected are: the Prevention stage of cancer control has not been addressed with intelligent algorithms, and the Early Detection stage has been very little addressed; private data sources could be beneficial in this type of research, but the difficulty in accessing them is a barrier for researchers. In addition, although Latin America and the Caribbean have a significant death rate from breast cancer, patients in this region have not been the subject of study and the participation of researchers on the subject has been almost nonexistent. Finally, there seems to be a great opportunity to generate proposals based on intelligent algorithms with low cost and time to implement that could directly impact patient survival, improving the health systems of the countries in the region.
Downloads
References
American Cancer Society, “What Is Cancer?,” www.cancer.org, Nov. 06, 2020. https://www.cancer.org/treatment/understanding-your-diagnosis/what-is-cancer.html.
J. Larry Jameson et al., Harrison’s principles of internal medicine Volume 1 [...], 20th ed. New York Chicago San Francisco Mcgraw Hill Education, 2018.
J. Ferlay, M. Ervik, F. Lam, M. Colombet, L. Mery, M. Piñeros, A. Znaor, I. Soerjomataram, F. Bray (2020). Global Cancer Observatory: Cancer Today. Lyon, France: International Agency for Research on Cancer, 2020. Available from: https://gco.iarc.fr/today, accessed [08 October 2022].
World Health Organization and Pan American Health Organization, “Breast Cancer,” Pan American Health Organization / World Health Organization. https://www3.paho.org/hq/index.php?option=com_content&view=article&id=5041:2011-breast-cancer&Itemid=3639&lang=en.
World Health Organization and Pan American Health Organization, “WHO outlines steps to save 7 million lives from cancer,” Pan American Health Organization / World Health Organization, Feb. 04, 2020. https://www3.paho.org/hq/index.php?option=com_content&view=article&id=15708.
U.S. Department of Health and Human Services, National Institutes of Health, and National Cancer Institute, “Breast Cancer Risk Assessment Tool,” Breast Cancer Risk Assessment Tool, 2019. https://bcrisktool.cancer.gov/calculator.html.
W. Scott Richardson, M. C. Wilson, J. Nishikawa, and R. S. A. Hayward, “The well-built clinical question: a key to evidence-based decisions,” ACP Journal Club, vol. 123, no. 3, p. A12, Nov. 1995, DOI: 10.7326/acpjc-1995-123-3-a12.
D. Moher, A. Liberati, J. Tetzlaff, and D. G. Altman, “Preferred Reporting Items for Systematic Reviews and Meta-Analyses: the PRISMA Statement,” PLoS Medicine, vol. 6, no. 7, p. e1000097, Jul. 2009, DOI: 10.1371/journal.pmed.1000097.
M. Hosni, I. Abnane, A. Idri, J. M. Carrillo de Gea, and J. L. Fernández Alemán, “Reviewing ensemble classification methods in breast cancer,” Computer Methods and Programs in Biomedicine, vol. 177, no. 1, pp. 89–112, Aug. 2019, DOI: 10.1016/j.cmpb.2019.05.019.
J. Li., Z. Zhou, J. Dong, Y. Fu, Y. Li, Z. Luan, X. Peng. “Predicting breast cancer 5-year survival using machine learning: A systematic review”. Plos One, vol 16, no. 4, pp. e0250370, April 2021, DOI: 10.1371/journal.pone.0250370.
N. I. R. Yassin, S. Omran, E. M. F. El Houby, and H. Allam, “Machine learning techniques for breast cancer computer aided diagnosis using different image modalities: A systematic review,” Computer Methods and Programs in Biomedicine, vol. 156, pp. 25–45, Mar. 2018, DOI: 10.1016/j.cmpb.2017.12.012.
World Health Organization, Cancer Control: Knowledge into Action: WHO Guide for Effective Programmes: Module 1: Planning. World Health Organization, 2006.
World Health Organization and Pan American Health Organization, “Prevention: Breast Cancer Risk Factors and Prevention - PAHO/WHO | Pan American Health Organization,” www.paho.org, Sep. 27, 2016. https://www.paho.org/en/documents/prevention-breast-cancer-risk-factors-and-prevention.
Pan American Health Organization, “Planning: Planning comprehensive breast cancer programs: call to action,” 2016. https://www.paho.org/hq/dmdocuments/2016/KNOWLEDGE-SUMMARY---PLANNING-2.pdf.
World Health Organization, “Breast cancer,” www.who.int, Mar. 26, 2021. https://www.who.int/news-room/fact-sheets/detail/breast-cancer.
D. Dua and C. Graff, “UCI Machine Learning Repository,” Machine Learning Repository, 2019. http://archive.ics.uci.edu/ml.
P. Henriques Abreu, M. Seoane Santos, M. Henriques Abreu, B. Andrade, and D. Castro Silva, “Predicting Breast Cancer Recurrence Using Machine Learning Techniques,” ACM Computing Surveys, vol. 49, no. 3, pp. 1–40, Oct. 2016, doi: 10.1145/2988544.
Y. Wang, Y. Gao, M. Battsend, K. Chen, W. Lu, and Y. Wang, “Development of a risk assessment tool for projecting individualized probabilities of developing breast cancer for Chinese women,” Tumor Biology, vol. 35, no. 11, pp. 10861–10869, Aug. 2014, doi: 10.1007/s13277-014-1967-0.
M. U. Salma and N. A. Doreswamy, “Hybrid BATGSA: a metaheuristic model for classification of breast cancer data,” International Journal of Advanced Intelligence Paradigms, vol. 15, no. 2, p. 207, 2020, DOI: 10.1504/ijaip.2020.105144.
V. J. Kadam, S. M. Jadhav, and K. Vijayakumar, “Breast Cancer Diagnosis Using Feature Ensemble Learning Based on Stacked Sparse Autoencoders and Softmax Regression,” Journal of Medical Systems, vol. 43, no. 8, Jul. 2019, DOI: 10.1007/s10916-019-1397-z.
P. D. Hung, T. D. Hanh, and V. T. Diep, “Breast Cancer Prediction Using Spark MLlib and ML Packages,” Proceedings of the 2018 5th International Conference on Bioinformatics Research and Applications, vol. 1, no. 1, Dec. 2018, DOI: 10.1145/3309129.3309133.
M. Pinto, O. Alkada, and H. Wei, “Health care AI: predicting breast cancer with machine learning,” Journal of Computing Sciences in Colleges, vol. 34, no. 2, pp. 65–71, Dec. 2018, DOI: abs/10.5555/3282588.3282598.
T. Gao et al., “Predicting pathological response to neoadjuvant chemotherapy in breast cancer patients based on imbalanced clinical data,” Personal and Ubiquitous Computing, vol. 22, no. 5–6, pp. 1039–1047, May 2018, DOI: 10.1007/s00779-018-1144-3.
T. K. Avramov and D. Si, “Comparison of Feature Reduction Methods and Machine Learning Models for Breast Cancer Diagnosis,” Proceedings of the International Conference on Compute and Data Analysis - ICCDA ’17, vol. 1, no. 1, pp. 69–74, May 2017, DOI: 10.1145/3093241.3093290.
I. Gómez, N. Ribelles, L. Franco, E. Alba, and J. M. Jerez, “Supervised discretization can discover risk groups in cancer survival analysis,” Computer Methods and Programs in Biomedicine, vol. 136, no. 1, pp. 11–19, Nov. 2016, DOI: 10.1016/j.cmpb.2016.08.006.
G. Napolitano, A. Marshall, P. Hamilton, and A. T. Gavin, “Machine learning classification of surgical pathology reports and chunk recognition for information extraction noise reduction,” Artificial Intelligence in Medicine, vol. 70, no. 1, pp. 77–83, Jun. 2016, DOI: 10.1016/j.artmed.2016.06.001.
S. Jhajharia, S. Verma, and R. Kumar, “Predictive Analytics for Breast Cancer Survivability,” Proceedings of the Second International Conference on Information and Communication Technology for Competitive Strategies - ICTCS ’16, vol. 1, no. 1, pp. 1–5, Mar. 2016, DOI: 10.1145/2905055.2905084.
M. Salehi, J. Razmara, and S. Lotfi, “A Novel Data Mining on Breast Cancer Survivability Using MLP Ensemble Learners,” The Computer Journal, vol. 63, no. 3, pp. 435–447, May 2019, DOI: 10.1093/comjnl/bxz051.
V. A. Telsang and K. Hegde, “Breast Cancer Prediction Analysis using Machine Learning Algorithms,” 2020 International Conference on Communication, Computing and Industry 4.0 (C2I4), vol. 1, no. 1, pp. 1–5, Dec. 2020, DOI: 10.1109/c2i451079.2020.9368911.
Y. Wu, “Diagnosis of Breast Cancer Based on Support Vector Machine and Random Forest Methods,” 2020 International Conference on Computing and Data Science (CDS), vol. 1, no. 1, pp. 147–151, Aug. 2020, DOI: 10.1109/cds49703.2020.00036.
A. Ganesan, G. Nagabushnam, A. Paul, and K. H. Kim, “Genetic Analysis for Breast Cancer Prediction and Diagnosis,” 2020 8th International Conference on Orange Technology (ICOT), vol. 1, no. 1, pp. 1–4, Dec. 2020, DOI: 10.1109/icot51877.2020.9468772.
M. R. Basunia et al., “On Predicting and Analyzing Breast Cancer using Data Mining Approach,” 2020 IEEE Region 10 Symposium (TENSYMP), vol. 1, no. 1, pp. 1257–1260, 2020, DOI: 10.1109/tensymp50017.2020.9230871.
S. J. Pasha and E. S. Mohamed, “Bio inspired Ensemble Feature Selection (BEFS) Model with Machine Learning and Data Mining Algorithms for Disease Risk Prediction,” 2019 5th International Conference On Computing, Communication, Control And Automation (ICCUBEA), vol. 1, no. 1, pp. 1–6, Sep. 2019, DOI: 10.1109/iccubea47591.2019.9129304.
M. U. Ghani, T. M. Alam, and F. H. Jaskani, “Comparison of Classification Models for Early Prediction of Breast Cancer,” 2019 International Conference on Innovative Computing (ICIC), vol. 1, no. 1, pp. 1–6, Nov. 2019, DOI: 10.1109/icic48496.2019.8966691.
A. Sinha, B. Sahoo, S. S. Rautaray, and M. Pandey, “Improved Framework for Breast Cancer Prediction Using Frequent Itemsets Mining for Attributes Filtering,” 2019 International Conference on Intelligent Computing and Control Systems (ICCS), vol. 1, no. 1, pp. 979–982, May 2019, DOI: 10.1109/iccs45141.2019.9065877.
S. Laghmati, A. Tmiri, and B. Cherradi, “Machine Learning based System for Prediction of Breast Cancer Severity,” 2019 International Conference on Wireless Networks and Mobile Communications (WINCOM), vol. 1, no. 1, pp. 1–5, Oct. 2019, DOI: 10.1109/wincom47513.2019.8942575.
P. Singhal and S. Pareek, “Artificial Neural Network for Prediction of Breast Cancer,” 2018 2nd International Conference on I-SMAC (IoT in Social, Mobile, Analytics and Cloud) (I-SMAC)I-SMAC (IoT in Social, Mobile, Analytics and Cloud) (I-SMAC), 2018 2nd International Conference on, vol. 1, no. 1, pp. 464–468, Aug. 2018, DOI: 10.1109/i-smac.2018.8653700.
X. Zhang and Y. Sun, “Breast cancer risk prediction model based on C5.0 algorithm for postmenopausal women,” 2018 International Conference on Security, Pattern Analysis, and Cybernetics (SPAC), vol. 1, no. 1, pp. 321–325, Dec. 2018, DOI: 10.1109/spac46244.2018.8965528.
M. H. Tafish and A. M. El-Halees, “Breast Cancer Severity Degree Predication Using Data Mining Techniques in the Gaza Strip,” 2018 International Conference on Promising Electronic Technologies (ICPET), vol. 1, no. 1, pp. 124–128, Oct. 2018, DOI: 10.1109/icpet.2018.00029.
S. B. Sakri, N. B. Abdul Rashid, and Z. Muhammad Zain, “Particle Swarm Optimization Feature Selection for Breast Cancer Recurrence Prediction,” IEEE Access, vol. 6, no. 1, pp. 29637–29647, Jun. 2018, DOI: 10.1109/access.2018.2843443.
U. Ojha and S. Goel, “A study on prediction of breast cancer recurrence using data mining techniques,” 2017 7th International Conference on Cloud Computing, Data Science & Engineering - Confluence, vol. 1, no. 1, pp. 527–530, Jan. 2017, DOI: 10.1109/confluence.2017.7943207.
S. G. Durai, S. H. Ganesh, and A. J. Christy, “Novel Linear Regressive Classifier for the Diagnosis of Breast Cancer,” 2017 World Congress on Computing and Communication Technologies (WCCCT), vol. 1, no. 1, pp. 136–139, Feb. 2017, DOI: 10.1109/wccct.2016.40.
S. Jhajharia, H. Kumar Varshney, S. Verma, and R. Kumar, “A neural network based breast cancer prognosis model with PCA processed features,” 2016 International Conference on Advances in Computing, Communications and Informatics (ICACCI), vol. 1, no. 1, pp. 1896–1901, Sep. 2016, DOI: 10.1109/ICACCI.2016.7732327.
A. I. Pritom, Md. A. R. Munshi, S. A. Sabab, and S. Shihab, “Predicting breast cancer recurrence using effective classification and feature selection technique,” 2016 19th International Conference on Computer and Information Technology (ICCIT), vol. 1, no. 1, pp. 310–314, Dec. 2016, DOI: 10.1109/iccitechn.2016.7860215.
M. Pellegrini, “Accurate prediction of breast cancer survival through coherent voting networks with gene expression profiling,” Scientific Reports, vol. 11, no. 1, pp. 14645 (2021), Jul. 2021, DOI: 10.1038/s41598-021-94243-z.
L. K. Tsou et al., “Comparative study between deep learning and QSAR classifications for TNBC inhibitors and novel GPCR agonist discovery,” Scientific Reports, vol. 10, no. 1, p. 16771, Oct. 2020, DOI: 10.1038/s41598-020-73681-1.
B.-J. Cho, K. M. Kim, S.-E. Bilegsaikhan, and Y. J. Suh, “Machine learning improves the prediction of febrile neutropenia in Korean inpatients undergoing chemotherapy for breast cancer,” Scientific Reports, vol. 10, no. 1, p. 14803, Sep. 2020, DOI: 10.1038/s41598-020-71927-6.
S. Roy, R. Kumar, V. Mittal, and D. Gupta, “Classification models for Invasive Ductal Carcinoma Progression, based on gene expression data-trained supervised machine learning,” Scientific Reports, vol. 10, no. 1, p. 4113, Mar. 2020, DOI: 10.1038/s41598-020-60740-w.
P. Ghosh, A. Karim, S. T. Atik, S. Afrin, and Mohd. Saifuzzaman, “Expert cancer model using supervised algorithms with a LASSO selection approach,” International Journal of Electrical and Computer Engineering (IJECE), vol. 11, no. 3, p. 2631, Jun. 2021, DOI: 10.11591/ijece.v11i3.pp2631-2639.
B. Kim, J. Kim, I. Lim, D. H. Kim, S. M. Lim, and S. Woo, “Machine Learning Model for Lymph Node Metastasis Prediction in Breast Cancer Using Random Forest Algorithm and Mitochondrial Metabolism Hub Genes,” Applied Sciences, vol. 11, no. 7, p. 2897, Mar. 2021, DOI: 10.3390/app11072897.
S. A. Diwani and Z. O. Yonah, “Holistic diagnosis tool for prediction of benign and malignant breast cancer using data mining techniques,” International Journal of Computing and Digital Systems, vol. 10, no. 1, pp. 417–432, Apr. 2021, DOI: 10.12785/ijcds/100141.
M. A. Jabbar, “Breast Cancer Data Classification Using Ensemble Machine Learning,” Engineering and Applied Science Research, vol. 48, no. 1, pp. 65–72, Jan. 2021, DOI: https://ph01.tci-thaijo.org/index.php/easr/article/view/234959.
N. F. Omran, S. F. Abd-el Ghany, H. Saleh, and A. Nabil, “Breast Cancer Identification from Patients’ Tweet Streaming Using Machine Learning Solution on Spark,” Complexity, vol. 2021, no. 1, pp. 1–12, Jan. 2021, DOI: 10.1155/2021/6653508.
S. Liu et al., “Survival Time Prediction of Breast Cancer Patients Using Feature Selection Algorithm Crystall,” IEEE Access, vol. 9, no. 1, pp. 24433–24445, Jan. 2021, DOI: 10.1109/access.2021.3054823.
M. Razu. Ahmed, A. Ali, J. Roy, S. Ahmed, and N. Ahmed, “Breast Cancer Risk Prediction based on Six Machine Learning Algorithms,” 2020 IEEE Asia-Pacific Conference on Computer Science and Data Engineering (CSDE), vol. 1, no. 1, pp. 1–5, Dec. 2020, DOI: 10.1109/csde50874.2020.9411572.
D. Kurz and C. Axenie, “PERFECTO: Prediction of Extended Response and Growth Functions for Estimating Chemotherapy Outcomes in Breast Cancer,” 2020 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), vol. 1, no. 1, pp. 609–614, Dec. 2020, DOI: 10.1109/bibm49941.2020.9313551.
X. Zhou et al., “A New Deep Convolutional Neural Network Model for Automated Breast Cancer Detection,” 2020 7th International Conference on Behavioural and Social Computing (BESC), vol. 1, no. 1, pp. 1–4, Nov. 2020, DOI: 10.1109/besc51023.2020.9348322.
A. Gupta, D. Kaushik, M. Garg, and A. Verma, “Machine Learning model for Breast Cancer Prediction,” 2020 Fourth International Conference on I-SMAC (IoT in Social, Mobile, Analytics and Cloud) (I-SMAC), vol. 1, no. 1, pp. 472–477, Oct. 2020, DOI: 10.1109/i-smac49090.2020.9243323.
S. Mojrian et al., “Hybrid Machine Learning Model of Extreme Learning Machine Radial basis function for Breast Cancer Detection and Diagnosis; a Multilayer Fuzzy Expert System,” IEEE Xplore, Oct. 14, 2020. https://ieeexplore.ieee.org/abstract/document/9140744 (accessed Apr. 15, 2021).
S. Laghmati, B. Cherradi, A. Tmiri, O. Daanouni, and S. Hamida, “Classification of Patients with Breast Cancer using Neighbourhood Component Analysis and Supervised Machine Learning Techniques,” 2020 3rd International Conference on Advanced Communication Technologies and Networking (CommNet), vol. 1, no. 1, pp. 1–6, Sep. 2020, DOI: 10.1109/commnet49926.2020.9199633.
K. Juneja and C. Rana, “An improved weighted decision tree approach for breast cancer prediction,” International Journal of Information Technology, vol. 12, no. 3, pp. 797–804, Apr. 2018, DOI: 10.1007/s41870-018-0184-2.
P. Sengar, M. Gaikwad, and A. Nagdive, “Comparative Study of Machine Learning Algorithms for Breast Cancer Prediction,” 2020 Third International Conference on Smart Systems and Inventive Technology (ICSSIT), vol. 1, no. 1, pp. 796–801, Aug. 2020, DOI: 10.1109/icssit48917.2020.9214267.
S. Kabiraj et al., “Breast Cancer Risk Prediction using XGBoost and Random Forest Algorithm,” 2020 11th International Conference on Computing, Communication and Networking Technologies (ICCCNT), vol. 1, no. 1, pp. 1–4, Jul. 2020, DOI: 10.1109/icccnt49239.2020.9225451.
M. Anitha, S. Gayathri, S. Nickolas, and M. S. Bhanu, “Feature Engineering based Automatic Breast Cancer Prediction,” 2020 Second International Conference on Inventive Research in Computing Applications (ICIRCA), vol. 1, no. 1, pp. 247–256, Jul. 2020, DOI: 10.1109/icirca48905.2020.9182855.
S. Prakash and K. Visakha, “Breast Cancer Malignancy Prediction Using Deep Learning Neural Networks,” 2020 Second International Conference on Inventive Research in Computing Applications (ICIRCA), vol. 1, no. 1, pp. 88–92, Jul. 2020, DOI: 10.1109/icirca48905.2020.9183378.
G. Murtaza, L. Shuib, A. W. A. Wahab, G. Mujtaba, and G. Raza, “Ensembled deep convolution neural network-based breast cancer classification with misclassification reduction algorithms,” Multimedia Tools and Applications, vol. 79, no. 25–26, pp. 18447–18479, Mar. 2020, DOI: 10.1007/s11042-020-08692-1.
S. Ray, A. AlGhamdi, A. AlGhamdi, K. Alshouiliy, and D. P. Agrawal, “Selecting Features for Breast Cancer Analysis and Prediction,” 2020 International Conference on Advances in Computing and Communication Engineering (ICACCE), vol. 1, no. 1, pp. 1–6, Jun. 2020, DOI: 10.1109/icacce49060.2020.9154919.
H. Hajiabadi, V. Babaiyan, D. Zabihzadeh, and M. Hajiabadi, “Combination of loss functions for robust breast cancer prediction,” Computers & Electrical Engineering, vol. 84, no. 1, p. 106624, Jun. 2020, DOI: 10.1016/j.compeleceng.2020.106624.
K. Uyar, U. Ilhan, A. Ilhan, and E. I. Iseri, “Breast Cancer Prediction Using Neuro-Fuzzy Systems,” 2020 7th International Conference on Electrical and Electronics Engineering (ICEEE), vol. 1, no. 1, pp. 328–332, Apr. 2020, DOI: 10.1109/iceee49618.2020.9102476.
M. Abdar et al., “A new nested ensemble technique for automated diagnosis of breast cancer,” Pattern Recognition Letters, vol. 132, no. 1, pp. 123–131, Apr. 2020, DOI: 10.1016/j.patrec.2018.11.004.
M. Mondal, R. Semwal, U. Raj, I. Aier, and P. K. Varadwaj, “An entropy-based classification of breast cancerous genes using microarray data,” Neural Computing and Applications, vol. 1, no. 1, pp. 2397–2404, Nov. 2018, DOI: 10.1007/s00521-018-3864-8.
M. Shalini and S. Radhika, “Machine Learning techniques for Prediction from various Breast Cancer Datasets,” IEEE Xplore, Feb. 27, 2020. https://ieeexplore.ieee.org/document/9167657 (accessed Jul. 09, 2021).
N. Papandrianos, E. Papageorgiou, A. Anagnostis, and A. Feleki, “A Deep-Learning Approach for Diagnosis of Metastatic Breast Cancer in Bones from Whole-Body Scans,” Applied Sciences, vol. 10, no. 3, p. 997, Feb. 2020, DOI: 10.3390/app10030997.
G. Battineni, N. Chintalapudi, and F. Amenta, “Performance analysis of different machine learning algorithms in breast cancer predictions,” EAI Endorsed Transactions on Pervasive Health and Technology, vol. 6, no. 23, p. 166010, Sep. 2020, DOI: 10.4108/eai.28-5-2020.166010.
Z. M. Zain et al., “Predicting breast cancer recurrence using principal component analysis as feature extraction: an unbiased comparative analysis,” International Journal of Advances in Intelligent Informatics, vol. 6, no. 3, p. 313, Nov. 2020, DOI: 10.26555/ijain.v6i3.462.
V. S et al., “Multi-modal prediction of breast cancer using particle swarm optimization with non-dominating sorting,” International Journal of Distributed Sensor Networks, vol. 16, no. 11, p. 155014772097150, Nov. 2020, DOI: 10.1177/1550147720971505.
S. Jain and P. Kumar, “Prediction of Breast Cancer Using Machine Learning,” Recent Patents on Computer Science, vol. 12, no. 1, Jun. 2019, DOI: 10.2174/2213275912666190617160834.
Z. Ceylan, “Diagnosis of Breast Cancer Using Improved Machine Learning Algorithms Based on Bayesian Optimization,” International Journal of Intelligent Systems and Applications in Engineering, vol. 8, no. 3, pp. 121–130, Sep. 2020, DOI: 10.18201/ijisae.2020363531.
M. Alfifi, M. Shady, S. Bataineh, and M. Mezher, “Enhanced Artificial Intelligence System for Diagnosing and Predicting Breast Cancer using Deep Learning,” International Journal of Advanced Computer Science and Applications, vol. 11, no. 7, pp. 498–513, 2020, DOI: 10.14569/ijacsa.2020.0110763.
S. H. Kassani, P. H. Kassani, M. J. Wesolowski, K. A. Schneider, and R. Deters, “Classification of Histopathological Biopsy Images Using Ensemble of Deep Learning Networks,” arXiv:1909.11870 [cs, eess], Sep. 2019, Accessed: Nov. 02, 2021. [Online]. Available: https://arxiv.org/abs/1909.11870.
R. Dhanya, I. R. Paul, S. S. Akula, M. Sivakumar, and J. J. Nair, “F-test feature selection in Stacking ensemble model for breast cancer prediction,” Procedia Computer Science, vol. 171, no. 1, pp. 1561–1570, 2020, DOI: 10.1016/j.procs.2020.04.167.
P. Gupta and S. Garg, “Breast Cancer Prediction using varying Parameters of Machine Learning Models,” Procedia Computer Science, vol. 171, no. 1, pp. 593–601, 2020, DOI: 10.1016/j.procs.2020.04.064.
K. Gupta and N. Chawla, “Analysis of Histopathological Images for Prediction of Breast Cancer Using Traditional Classifiers with Pre-Trained CNN,” Procedia Computer Science, vol. 167, no. 1, pp. 878–889, 2020, DOI: 10.1016/j.procs.2020.03.427.
R. A. I. Alhayali, M. A. Ahmed, Y. M. Mohialden, and A. H. Ali, “Efficient method for breast cancer classification based on ensemble hoffeding tree and naïve Bayes,” Indonesian Journal of Electrical Engineering and Computer Science, vol. 18, no. 2, p. 1074, May 2020, DOI: 10.11591/ijeecs.v18.i2.pp1074-1080.
B. Al-Shargabi and F. Al-Shami, “An experimental study for breast cancer prediction algorithms,” Proceedings of the Second International Conference on Data Science, E-Learning and Information Systems - DATA ’19, vol. 1, no. 1, pp. 1–6, Dec. 2019, DOI: 10.1145/3368691.3368703.
K. Chakradeo, S. Vyawahare, and P. Pawar, “Breast Cancer Recurrence Prediction using Machine Learning,” 2019 IEEE Conference on Information and Communication Technology, pp. 1–7, Dec. 2019, DOI: 10.1109/cict48419.2019.9066248.
W. N. L. W. H. Ibeni, M. Z. M. Salikon, A. Mustapha, S. A. Daud, and M. N. M. Salleh, “Comparative analysis on bayesian classification for breast cancer problem,” Bulletin of Electrical Engineering and Informatics, vol. 8, no. 4, pp. 1303–1311, Dec. 2019, DOI: 10.11591/eei.v8i4.1628.
M. J. Domingo, B. D. Gerardo, and R. P. Medina, “Fuzzy decision tree for breast cancer prediction,” Proceedings of the International Conference on Advanced Information Science and System, vol. 1, no. 1, pp. 1–6, Nov. 2019, DOI: 10.1145/3373477.3373489.
T. Shouket, S. Mahmood, M. T. Hassan, and A. Iftikhar, “Overall and Disease-Free Survival Prediction of Postoperative Breast Cancer Patients using Machine Learning Techniques,” 2019 22nd International Multitopic Conference (INMIC), vol. 1, pp. 1–6, Nov. 2019, DOI: 10.1109/inmic48123.2019.9022756.
J. Akinsola, “Breast Cancer Predictive Analytics Using Supervised Machine Learning Techniques,” International Journal of Advanced Trends in Computer Science and Engineering, vol. 8, no. 6, pp. 3095–3104, Dec. 2019, DOI: 10.30534/ijatcse/2019/70862019.
G. Hungilo, G. Emmanuel, and A. W. R. Emanuel, “Performance Evaluation of Ensembles Algorithms in Prediction of Breast Cancer,” 2019 International Biomedical Instrumentation and Technology Conference (IBITeC), vol. 1, pp. 74–79, Oct. 2019, DOI: 10.1109/ibitec46597.2019.9091718.
J. Awatramani and N. Hasteer, “Early Stage Detection of Malignant Cells: A Step Towards Better Life,” 2019 International Conference on Computing, Communication, and Intelligent Systems (ICCCIS), vol. 1, pp. 262–267, Oct. 2019, DOI: 10.1109/icccis48478.2019.8974543.
A. K. Arslan, S. Yasar, and C. Colak, “Breast cancer classification using a constructed convolutional neural network on the basis of the histopathological images by an interactive web-based interface,” 2019 3rd International Symposium on Multidisciplinary Studies and Innovative Technologies (ISMSIT), vol. 1, no. 1, pp. 1–5, Oct. 2019, DOI: 10.1109/ismsit.2019.8932942.
K. Alshouiliy, A. Shivanna, S. Ray, A. AlGhamdi, and D. P. Agrawal, “Analysis and Prediction of Breast Cancer using AzureML Platform,” 2019 IEEE 10th Annual Information Technology, Electronics and Mobile Communication Conference (IEMCON), vol. 1, no. 1, pp. 0212–0218, Oct. 2019, DOI: 10.1109/iemcon.2019.8936294.
S. V. J. Jaikrishnan, O. Chantarakasemchit, and P. Meesad, “A Breakup Machine Learning Approach for Breast Cancer Prediction,” 2019 11th International Conference on Information Technology and Electrical Engineering (ICITEE), vol. 1, no. 1, pp. 1–6, Oct. 2019, DOI: 10.1109/iciteed.2019.8929977.
T. Brito-Sarracino, M. Rocha dos Santos, E. Freire Antunes, I. Batista de Andrade Santos, J. Coelho Kasmanas, and A. C. Ponce de Leon Ferreira de Carvalho, “Explainable Machine Learning for Breast Cancer Diagnosis,” 2019 8th Brazilian Conference on Intelligent Systems (BRACIS), vol. 1, no. 1, pp. 681–686, Oct. 2019, DOI: 10.1109/bracis.2019.00124.
S. Poorani and P. Balasubramanie, “Deep Neural Network Classifier in Breast Cancer Prediction,” International Journal of Engineering and Advanced Technology, vol. 9, no. 1, pp. 2106–2109, Oct. 2019, DOI: 10.35940/ijeat.a9664.109119.
F. Teixeira, J. L. Z. Montenegro, C. A. da Costa, and R. da Rosa Righi, “An Analysis of Machine Learning Classifiers in Breast Cancer Diagnosis,” 2019 XLV Latin American Computing Conference (CLEI), vol. 1, no. 1, pp. 1–10, Sep. 2019, DOI: 10.1109/clei47609.2019.235094.
S. Das and D. Biswas, “Prediction of Breast Cancer Using Ensemble Learning,” 2019 5th International Conference on Advances in Electrical Engineering (ICAEE), vol. 1, no. 1, pp. 804–808, Sep. 2019, DOI: 10.1109/icaee48663.2019.8975544.
T. I. Rohan, A. U. Rahman, A. B. Siddik, M. Islam, and Md. S. U. Yusuf, “A Precise Breast Cancer Detection Approach Using Ensemble of Random Forest with AdaBoost,” 2019 International Conference on Computer, Communication, Chemical, Materials and Electronic Engineering (IC4ME2), vol. 1, no. 1, pp. 1–4, Jul. 2019, DOI: 10.1109/ic4me247184.2019.9036697.
F. Y. A’la, A. E. Permanasari, and N. A. Setiawan, “A Comparative Analysis of Tree-based Machine Learning Algorithms for Breast Cancer Detection,” 2019 12th International Conference on Information & Communication Technology and System (ICTS), vol. 1, no. 1, pp. 55–59, Jul. 2019, DOI: 10.1109/ICTS.2019.8850975.
R. Dhanya, I. R. Paul, S. Sindhu Akula, M. Sivakumar, and J. J. Nair, “A Comparative Study for Breast Cancer Prediction using Machine Learning and Feature Selection,” 2019 International Conference on Intelligent Computing and Control Systems (ICCS), vol. 1, no. 1, pp. 1049–1055, May 2019, DOI: 10.1109/iccs45141.2019.9065563.
Naveen, R. K. Sharma, and A. Ramachandran Nair, “Efficient Breast Cancer Prediction Using Ensemble Machine Learning Models,” 2019 4th International Conference on Recent Trends on Electronics, Information, Communication & Technology (RTEICT), vol. 1, no. 1, pp. 100–104, May 2019, DOI: 10.1109/rteict46194.2019.9016968.
Harshitha, V. Chaitanya, S. M. Killedar, D. Revankar, and M. S. Pushpa, “Recognition and Prediction of Breast Cancer using Supervised Diagnosis,” 2019 4th International Conference on Recent Trends on Electronics, Information, Communication & Technology (RTEICT), vol. 1, no. 1, pp. 1436–1441, May 2019, DOI: 10.1109/rteict46194.2019.9016921.
A. M. Romano and A. A. Hernandez, “Enhanced Deep Learning Approach for Predicting Invasive Ductal Carcinoma from Histopathology Images,” 2019 2nd International Conference on Artificial Intelligence and Big Data (ICAIBD), vol. 1, no. 1, pp. 142–148, May 2019, DOI: 10.1109/icaibd.2019.8837044.
E. A. Bayrak, P. Kirci, and T. Ensari, “Comparison of Machine Learning Methods for Breast Cancer Diagnosis,” 2019 Scientific Meeting on Electrical-Electronics & Biomedical Engineering and Computer Science (EBBT), vol. 1, no. 1, pp. 1–3, Apr. 2019, DOI: 10.1109/ebbt.2019.8741990.
M. Vazifehdan, M. H. Moattar, and M. Jalali, “A hybrid Bayesian network and tensor factorization approach for missing value imputation to improve breast cancer recurrence prediction,” Journal of King Saud University - Computer and Information Sciences, vol. 31, no. 2, pp. 175–184, Apr. 2019, DOI: 10.1016/j.jksuci.2018.01.002.
T. Chandrasegar and S. B. Nikhilesh Vutukuri, “Optimized machine learning model using Decision Tree for cancer prediction,” 2019 Innovations in Power and Advanced Computing Technologies (i-PACT), vol. 1, no. 1, pp. 1–4, Mar. 2019, DOI: 10.1109/i-pact44901.2019.8960129.
B. Dai, R.-C. Chen, S.-Z. Zhu, and W.-W. Zhang, “Using Random Forest Algorithm for Breast Cancer Diagnosis,” 2018 International Symposium on Computer, Consumer and Control (IS3C), vol. 1, no. 1, pp. 449–452, Dec. 2018, DOI: 10.1109/is3c.2018.00119.
S. Bharati, M. A. Rahman, and P. Podder, “Breast Cancer Prediction Applying Different Classification Algorithm with Comparative Analysis using WEKA,” 2018 4th International Conference on Electrical Engineering and Information & Communication Technology (iCEEiCT), vol. 1, no. 1, pp. 581–584, Sep. 2018, DOI: 10.1109/ceeict.2018.8628084.
Y. Khourdifi and M. Bahaj, “Feature Selection with Fast Correlation-Based Filter for Breast Cancer Prediction and Classification Using Machine Learning Algorithms,” 2018 International Symposium on Advanced Electrical and Communication Technologies (ISAECT), vol. 1, no. 1, pp. 1–6, Nov. 2018, DOI: 10.1109/isaect.2018.8618688.
M. Karim, G. Wicaksono, I. G. Costa, S. Decker, and O. Beyan, “Prognostically Relevant Subtypes and Survival Prediction for Breast Cancer Based on Multimodal Genomics Data,” IEEE Access, vol. 7, no. 1, pp. 133850–133864, Sep. 2019, DOI: 10.1109/access.2019.2941796.
I. Mihaylov, M. Nisheva, and D. Vassilev, “Application of Machine Learning Models for Survival Prognosis in Breast Cancer Studies,” Information, vol. 10, no. 3, p. 93, Mar. 2019, DOI: 10.3390/info10030093.
V. Sharma, R. K. Rajasekaran, and S. Badhrinarayanan, “Visualization of Data Mining Techniques for the Prediction of Breast Cancer with High Accuracy Rates,” Journal of Computer Science, vol. 15, no. 1, pp. 118–130, Jan. 2019, DOI: 10.3844/jcssp.2019.118.130.
J. Xu, P. Wu, Y. Chen, and L. Zhang, “Comparison of Different Classification Methods for Breast Cancer Subtypes Prediction,” 2018 International Conference on Security, Pattern Analysis, and Cybernetics (SPAC), vol. 1, no. 1, pp. 91–96, Dec. 2018, DOI: 10.1109/spac46244.2018.8965553.
S. Sharma, A. Aggarwal, and T. Choudhury, “Breast Cancer Detection Using Machine Learning Algorithms,” 2018 International Conference on Computational Techniques, Electronics and Mechanical Systems (CTEMS), vol. 1, no. 1, pp. 114–118, Dec. 2018, DOI: 10.1109/ctems.2018.8769187.
S. N. Singh and S. Thakral, “Using Data Mining Tools for Breast Cancer Prediction and Analysis,” 2018 4th International Conference on Computing Communication and Automation (ICCCA), vol. 1, no. 1, pp. 1–4, Dec. 2018, DOI: 10.1109/ccaa.2018.8777713.
M. Gupta and B. Gupta, “An Ensemble Model for Breast Cancer Prediction Using Sequential Least Squares Programming Method (SLSQP),” 2018 Eleventh International Conference on Contemporary Computing (IC3), vol. 1, no. 1, pp. 1–3, Aug. 2018, DOI: 10.1109/ic3.2018.8530572.
P. Chauhan and A. Swami, “Breast Cancer Prediction Using Genetic Algorithm Based Ensemble Approach,” 2018 9th International Conference on Computing, Communication and Networking Technologies (ICCCNT), vol. 1, no. 1, pp. 1–8, Jul. 2018, DOI: 10.1109/icccnt.2018.8493927.
A. Bharat, N. Pooja, and R. A. Reddy, “Using Machine Learning algorithms for breast cancer risk prediction and diagnosis,” 2018 3rd International Conference on Circuits, Control, Communication and Computing (I4C), vol. 1, no. 1, pp. 1–4, Oct. 2018, DOI: 10.1109/cimca.2018.8739696.
S. Murugan, B. M. Kumar, and S. Amudha, “Classification and Prediction of Breast Cancer using Linear Regression, Decision Tree and Random Forest,” 2017 International Conference on Current Trends in Computer, Electrical, Electronics and Communication (CTCEEC), vol. 1, no. 1, pp. 763–766, Sep. 2017, DOI: 10.1109/ctceec.2017.8455058.
D. Rafi and C. Bharathi, “A case study approach for Brest cancer prediction using feature selection method based on AOC and SVM,” Advances in Modelling and Analysis B, vol. 61, no. 3, pp. 145–150, Sep. 2018, DOI: 10.18280/ama_b.610307.
M. Liang, L. Huang, and W. Ahmad, “Breast Cancer Intelligent Diagnosis Based on Subtractive Clustering Adaptive Neural Fuzzy Inference System and Information Gain,” 2017 International Conference on Computer Systems, Electronics and Control (ICCSEC), vol. 1, no. 1, pp. 152–156, Dec. 2017, DOI: 10.1109/iccsec.2017.8446831.
A. Sethi, “Analogizing of Evolutionary and Machine Learning Algorithms for Prognosis of Breast Cancer,” 2018 7th International Conference on Reliability, Infocom Technologies and Optimization (Trends and Future Directions) (ICRITO), vol. 1, no. 1, pp. 252–255, Aug. 2018, DOI: 10.1109/icrito.2018.8748502.
M. Islam, H. Iqbal, Md. R. Haque, and Md. K. Hasan, “Prediction of breast cancer using support vector machine and K-Nearest neighbors,” 2017 IEEE Region 10 Humanitarian Technology Conference (R10-HTC), vol. 1, no. 1, pp. 226–229, Dec. 2017, DOI: 10.1109/r10-htc.2017.8288944.
T. Badriyah, R. Fauzyah, I. Syarif, and P. Kristalina, “Mobile personal health record (mPHR) for Breast Cancer using prediction modeling,” 2017 Second International Conference on Informatics and Computing (ICIC), no. ``, pp. 1–4, Nov. 2017, DOI: 10.1109/iac.2017.8280639.
B. K. Singh, K. Verma, L. Panigrahi, and A. S. Thoke, “Integrating radiologist feedback with computer aided diagnostic systems for breast cancer risk prediction in ultrasonic images: An experimental investigation in machine learning paradigm,” Expert Systems with Applications, vol. 90, no. 1, pp. 209–223, Dec. 2017, DOI: 10.1016/j.eswa.2017.08.020.
V. F. Adegoke, D. Chen, E. Banissi, and S. Barikzai, “Prediction of breast cancer survivability using ensemble algorithms,” 2017 International Conference on Smart Systems and Technologies (SST), vol. 1, no. 1, pp. 223–231, Oct. 2017, DOI: 10.1109/sst.2017.8188699.
H. Ponce and M. de Lourdes Martinez-Villasenor, “Interpretability of artificial hydrocarbon networks for breast cancer classification,” 2017 International Joint Conference on Neural Networks (IJCNN), vol. 1, no. 1, pp. 3535–3542, May 2017, DOI: 10.1109/ijcnn.2017.7966301.
J. Diz, G. Marreiros, and A. Freitas, “Applying Data Mining Techniques to Improve Breast Cancer Diagnosis,” Journal of Medical Systems, vol. 40, no. 9, Aug. 2016, DOI: 10.1007/s10916-016-0561-y.
D. Kaushik and K. Kaur, “Application of Data Mining for high accuracy prediction of breast tissue biopsy results,” 2016 Third International Conference on Digital Information Processing, Data Mining, and Wireless Communications (DIPDMWC), vol. 1, no. 1, pp. 40–45, Jul. 2016, DOI: 10.1109/dipdmwc.2016.7529361.
S. Muthuselvan, K. S. Sundaram, and Prabasheela, “Prediction of breast cancer using classification rule mining techniques in blood test datasets,” 2016 International Conference on Information Communication and Embedded Systems (ICICES), vol. 1, no. 1, pp. 1–5, Feb. 2016, DOI: 10.1109/icices.2016.7518932.
H. Asri, H. Mousannif, H. A. Moatassime, and T. Noel, “Using Machine Learning Algorithms for Breast Cancer Risk Prediction and Diagnosis,” Procedia Computer Science, vol. 83, no. 1, pp. 1064–1069, 2016, DOI: 10.1016/j.procs.2016.04.224.
V. Chaurasia and S. Pal, “Stacking-Based Ensemble Framework and Feature Selection Technique for the Detection of Breast Cancer,” SN Computer Science, vol. 2, no. 2, p. 67, Feb. 2021, DOI: 10.1007/s42979-021-00465-3.
M. Islam, Md. R. Haque, H. Iqbal, Md. M. Hasan, M. Hasan, and M. N. Kabir, “Breast Cancer Prediction: A Comparative Study Using Machine Learning Techniques,” SN Computer Science, vol. 1, no. 5, p. 290, Sep. 2020, DOI: 10.1007/s42979-020-00305-w.
V. Chaurasia and S. Pal, “Applications of Machine Learning Techniques to Predict Diagnostic Breast Cancer,” SN Computer Science, vol. 1, no. 5, p. 270, Aug. 2020, DOI: 10.1007/s42979-020-00296-8.
J. M. Valencia-Moreno, E. G. López, J. F. R. Pérez, J. P. F. Rodríguez, and O. Á. Xochihua, “Exploring Breast Cancer Prediction for Cuban Women,” Advances in Intelligent Systems and Computing, vol. 1, no. 1, pp. 480–489, Jan. 2020, DOI: 10.1007/978-3-030-40690-5_47.
S. A. Mohammed, S. Darrab, S. A. Noaman, and G. Saake, “Analysis of Breast Cancer Detection Using Different Machine Learning Techniques,” Data Mining and Big Data, vol. 1, no. 1, pp. 108–117, 2020, DOI: 10.1007/978-981-15-7205-0_10.
Y. Huang et al., “Prediction of Tumor Shrinkage Pattern to Neoadjuvant Chemotherapy Using a Multiparametric MRI-Based Machine Learning Model in Patients With Breast Cancer,” Frontiers in Bioengineering and Biotechnology, vol. 9, no. 1, pp. 1–15, Jul. 2021, DOI: 10.3389/fbioe.2021.662749.
H. Moghadas-Dastjerdi et al., “Prediction of chemotherapy response in breast cancer patients at pre-treatment using second derivative texture of CT images and machine learning,” Translational Oncology, vol. 14, no. 10, p. 101183, Oct. 2021, DOI: 10.1016/j.tranon.2021.101183.
Y. Dou and W. Meng, “An Optimization Algorithm for Computer-Aided Diagnosis of Breast Cancer Based on Support Vector Machine,” Frontiers in Bioengineering and Biotechnology, vol. 9, no. 1, p. 581, Jul. 2021, DOI: 10.3389/fbioe.2021.698390.
C. Kolios et al., “MRI texture features from tumor core and margin in the prediction of response to neoadjuvant chemotherapy in patients with locally advanced breast cancer,” Oncotarget, vol. 1, no. 1, pp. 1354–1365, Jan. 2021, DOI: 10.18632/oncotarget.28002.
Q. Li, Q. Xiao, J. Li, Z. Wang, H. Wang, and Y. Gu, “Value of Machine Learning with Multiphases CE-MRI Radiomics for Early Prediction of Pathological Complete Response to Neoadjuvant Therapy in HER2-Positive Invasive Breast Cancer,” Cancer Management and Research, vol. Volume 13, no. 1, pp. 5053–5062, Jun. 2021, DOI: 10.2147/cmar.s304547.
Y. Yu et al., “Magnetic resonance imaging radiomics predicts preoperative axillary lymph node metastasis to support surgical decisions and is associated with tumor microenvironment in invasive breast cancer: A machine learning, multicenter study,” EBioMedicine, vol. 69, no. 1, p. 103460, Jul. 2021, DOI: 10.1016/j.ebiom.2021.103460.
L. Hussain et al., “Machine learning classification of texture features of MRI breast tumor and peri-tumor of combined pre- and early treatment predicts pathologic complete response,” BioMedical Engineering, vol. 20, no. 1, p. 63, Jun. 2021, DOI: 10.1186/s12938-021-00899-z.
C. Kuo, H. Wang, and L. Tseng, “Using data mining technology to predict medication‐taking behaviour in women with breast cancer: A retrospective study,” Nursing Open, vol. 1, no. 1, pp. 1–11, Jun. 2021, DOI: 10.1002/nop2.963.
H. Xu et al., “Multi-Omics Marker Analysis Enables Early Prediction of Breast Tumor Progression,” Frontiers in Genetics, vol. 12, no. 1, p. 803, Jun. 2021, DOI: 10.3389/fgene.2021.670749.
B. O. Macaulay, B. S. Aribisala, S. A. Akande, B. A. Akinnuwesi, and O. A. Olabanjo, “Breast cancer risk prediction in African women using Random Forest Classifier,” Cancer Treatment and Research Communications, vol. 28, no. 1, p. 100396, May 2021, DOI: 10.1016/j.ctarc.2021.100396.
J. Kim et al., “Deep Learning-Based Prediction Model for Breast Cancer Recurrence Using Adjuvant Breast Cancer Cohort in Tertiary Cancer Center Registry,” Frontiers in Oncology, vol. 11, no. 1, May 2021, DOI: 10.3389/fonc.2021.596364.
J. Sanyal, A. Tariq, A. W. Kurian, D. Rubin, and I. Banerjee, “Weakly supervised temporal model for prediction of breast cancer distant recurrence,” Scientific Reports, vol. 11, no. 1, pp. 1–11, May 2021, DOI: 10.1038/s41598-021-89033-6.
M. Panagopoulou, M. Karaglani, V. G. Manolopoulos, I. Iliopoulos, I. Tsamardinos, and E. Chatzaki, “Deciphering the Methylation Landscape in Breast Cancer: Diagnostic and Prognostic Biosignatures through Automated Machine Learning,” Cancers, vol. 13, no. 7, p. 1677, Apr. 2021, DOI: 10.3390/cancers13071677.
N. Bakx, H. Bluemink, E. Hagelaar, M. van der Sangen, J. Theuws, and C. Hurkmans, “Development and evaluation of radiotherapy deep learning dose prediction models for breast cancer,” Physics and Imaging in Radiation Oncology, vol. 17, no. 1, pp. 65–70, Jan. 2021, DOI: 10.1016/j.phro.2021.01.006.
S. Abbas et al., “BCD-WERT: a novel approach for breast cancer detection using whale optimization based efficient features and extremely randomized tree algorithm,” PeerJ Computer Science, vol. 7, no. 1, p. e390, Mar. 2021, DOI: 10.7717/peerj-cs.390.
R. Massafra et al., “A Clinical Decision Support System for Predicting Invasive Breast Cancer Recurrence: Preliminary Results,” Frontiers in Oncology, vol. 11, no. 1, p. 284, Mar. 2021, DOI: 10.3389/fonc.2021.576007.
S. Nguyen et al., “Preoperative Prediction of Lymph Node Metastasis from Clinical DCE MRI of the Primary Breast Tumor Using a 4D CNN,” Medical Image Computing and Computer Assisted Intervention – MICCAI 2020, vol. 1, no. 1, pp. 326–334, Sep. 2020, DOI: 10.1007/978-3-030-59713-9_32.
R. Kothari et al., “Raman spectroscopy and artificial intelligence to predict the Bayesian probability of breast cancer,” Scientific Reports, vol. 11, no. 1, pp. 1–17, Mar. 2021, DOI: 10.1038/s41598-021-85758-6.
W. Tao et al., “Machine Learning Based on Multi-Parametric MRI to Predict Risk of Breast Cancer,” Frontiers in Oncology, vol. 11, no. 1, p. 570747, Feb. 2021, DOI: 10.3389/fonc.2021.570747.
H. Chereda et al., “Explaining decisions of graph convolutional neural networks: patient-specific molecular subnetworks responsible for metastasis prediction in breast cancer,” Genome Medicine, vol. 13, no. 1, pp. 1–16, Mar. 2021, DOI: 10.1186/s13073-021-00845-7.
Z. He, J. Zhang, X. Yuan, and Y. Zhang, “Integrating Somatic Mutations for Breast Cancer Survival Prediction Using Machine Learning Methods,” Frontiers in Genetics, vol. 11, no. 1, pp. 1-12, Jan. 2021, DOI: 10.3389/fgene.2020.632901.
J. Wu and C. Hicks, “Breast Cancer Type Classification Using Machine Learning,” Journal of Personalized Medicine, vol. 11, no. 2, p. 61, Jan. 2021, DOI: 10.3390/jpm11020061.
N. Al-Azzam and I. Shatnawi, “Comparing supervised and semi-supervised Machine Learning Models on Diagnosing Breast Cancer,” Annals of Medicine and Surgery, vol. 62, no. 1, pp. 53–64, Feb. 2021, DOI: 10.1016/j.amsu.2020.12.043.
H. Yazici et al., “New Approach for Risk Estimation Algorithms of BRCA1/2 Negativeness Detection with Modelling Supervised Machine Learning Techniques,” Disease Markers, vol. 2020, no. 1, pp. 1–7, Dec. 2020, DOI: 10.1155/2020/8594090.
O. Nave and M. Elbaz, “Artificial immune system features added to breast cancer clinical data for machine learning (ML) applications,” Biosystems, vol. 1, no. 1, p. 104341, Jan. 2021, DOI: 10.1016/j.biosystems.2020.104341.
D. Song, X. Man, M. Jin, Q. Li, H. Wang, and Y. Du, “A Decision-Making Supporting Prediction Method for Breast Cancer Neoadjuvant Chemotherapy,” Frontiers in Oncology, vol. 10, no. 1, p. 2879, Jan. 2021, DOI: 10.3389/fonc.2020.592556.
H. Yang, Y. Wang, H. Peng, and C. Huang, “Breath biopsy of breast cancer using sensor array signals and machine learning analysis,” Scientific Reports, vol. 11, no. 1, Jan. 2021, DOI: 10.1038/s41598-020-80570-0.
S. J. Lou et al., “Machine Learning Algorithms to Predict Recurrence within 10 Years after Breast Cancer Surgery: A Prospective Cohort Study,” Cancers, vol. 12, no. 12, p. 3817, Dec. 2020, DOI: 10.3390/cancers12123817.
K. Dembrower et al., “Effect of artificial intelligence-based triaging of breast cancer screening mammograms on cancer detection and radiologist workload: a retrospective simulation study,” The Lancet Digital Health, vol. 2, no. 9, pp. e468–e474, Sep. 2020, DOI: 10.1016/S2589-7500(20)30185-0.
F. Ben Azzouz et al., “Development of an absolute assignment predictor for triple-negative breast cancer subtyping using machine learning approaches,” Computers in Biology and Medicine, vol. 129, no. 1, p. 104171, Feb. 2021, DOI: 10.1016/j.compbiomed.2020.104171.
H. Wang, Y. Li, S. A. Khan, and Y. Luo, “Prediction of breast cancer distant recurrence using natural language processing and knowledge-guided convolutional neural network,” Artificial Intelligence in Medicine, vol. 110, no. 1, p. 101977, Nov. 2020, DOI: 10.1016/j.artmed.2020.101977.
V. C.-H. Chen, T.-Y. Lin, D.-C. Yeh, J.-W. Chai, and J.-C. Weng, “Functional and Structural Connectome Features for Machine Learning Chemo-Brain Prediction in Women Treated for Breast Cancer with Chemotherapy,” Brain Sciences, vol. 10, no. 11, p. 851, Nov. 2020, DOI: 10.3390/brainsci10110851.
Z. Yu, Z. Wang, X. Yu, and Z. Zhang, “RNA-Seq-Based Breast Cancer Subtypes Classification Using Machine Learning Approaches,” Computational Intelligence and Neuroscience, vol. 2020, no. 1, pp. 1–13, Oct. 2020, DOI: 10.1155/2020/4737969.
X. Zhong et al., “Multidimensional Machine Learning Personalized Prognostic Model in an Early Invasive Breast Cancer Population-Based Cohort in China: Algorithm Validation Study,” JMIR Medical Informatics, vol. 8, no. 11, p. e19069, Nov. 2020, DOI: 10.2196/19069.
A. Dasgupta et al., “Quantitative ultrasound radiomics using texture derivatives in prediction of treatment response to neo-adjuvant chemotherapy for locally advanced breast cancer,” Oncotarget, vol. 11, no. 42, pp. 3782–3792, Oct. 2020, DOI: 10.18632/oncotarget.27742.
D. Arefan, R. Chai, M. Sun, M. L. Zuley, and S. Wu, “Machine learning prediction of axillary lymph node metastasis in breast cancer: 2D versus 3D radiomic features,” Medical Physics, vol. 1, pp. 6334–6342, Nov. 2020, DOI: 10.1002/mp.14538.
A. Mosayebi, B. Mojaradi, A. B. Naeini, and S. H. K. Hosseini, “Modeling and comparing data mining algorithms for prediction of recurrence of breast cancer.,” PLoS ONE, vol. 15, no. 10, pp. e0237658–e0237658, Oct. 2020, DOI: 10.1371/journal.pone.0237658.
S. Chen et al., “Machine Learning-Based Radiomics Nomogram Using Magnetic Resonance Images for Prediction of Neoadjuvant Chemotherapy Efficacy in Breast Cancer Patients,” Frontiers in Oncology, vol. 10, p. 1410, Aug. 2020, DOI: 10.3389/fonc.2020.01410.
Y. Hao et al., “Improving Model Performance on the Stratification of Breast Cancer Patients by Integrating Multiscale Genomic Features,” BioMed Research International, vol. 2020, no. 1, pp. 1–12, Aug. 2020, DOI: 10.1155/2020/1475368.
S. T. Kakileti, G. Manjunath, A. Dekker, and L. Wee, “Robust Estimation of Breast Cancer Incidence Risk in Presence of Incomplete or Inaccurate Information,” Asian Pacific Journal of Cancer Prevention, vol. 21, no. 8, pp. 2307–2313, Aug. 2020, DOI: 10.31557/apjcp.2020.21.8.2307.
D. Gu, K. Su, and H. Zhao, “A case-based ensemble learning system for explainable breast cancer recurrence prediction,” Artificial Intelligence in Medicine, vol. 1, no. 1, p. 101858, Jun. 2020, DOI: 10.1016/j.artmed.2020.101858.
T. A’mar et al., “Incorporating Breast Cancer Recurrence Events Into Population-Based Cancer Registries Using Medical Claims: Cohort Study,” JMIR Cancer, vol. 6, no. 2, p. e18143, Aug. 2020, DOI: 10.2196/18143.
H. Rahbar et al., “The Value of Patient and Tumor Factors in Predicting Preoperative Breast MRI Outcomes,” Radiology: Imaging Cancer, vol. 2, no. 4, p. e190099, Jul. 2020, DOI: 10.1148/rycan.2020190099.
X. Zhuang et al., “Multiparametric MRI-based radiomics analysis for the prediction of breast tumor regression patterns after neoadjuvant chemotherapy,” Translational Oncology, vol. 13, no. 11, p. 100831, Nov. 2020, DOI: 10.1016/j.tranon.2020.100831.
H. Behravan, J. M. Hartikainen, M. Tengström, V. Kosma, and A. Mannermaa, “Predicting breast cancer risk using interacting genetic and demographic factors and machine learning,” Scientific Reports, vol. 10, no. 1, pp. 1–16, Jul. 2020, DOI: 10.1038/s41598-020-66907-9.
H. Moghadas-Dastjerdi, H. R. Sha-E-Tallat, L. Sannachi, A. Sadeghi-Naini, and G. J. Czarnota, “A priori prediction of tumour response to neoadjuvant chemotherapy in breast cancer patients using quantitative CT and machine learning,” Scientific Reports, vol. 10, no. 1, pp. 1–11, Jul. 2020, DOI: 10.1038/s41598-020-67823-8.
T. Jansen, G. Geleijnse, M. Van Maaren, M. P. Hendriks, A. Ten Teije, and A. Moncada-Torres, “Machine Learning Explainability in Breast Cancer Survival,” Digital Personalized Health and Medicine, vol. 1, pp. 307–311, 2020, DOI: 10.3233/SHTI200172.
C. Ming, V. Viassolo, N. Probst-Hensch, I. D. Dinov, P. O. Chappuis, and M. C. Katapodi, “Machine learning-based lifetime breast cancer risk reclassification compared with the BOADICEA model: impact on screening recommendations,” British Journal of Cancer, vol. 123, no. 5, pp. 860–867, Jun. 2020, DOI: 10.1038/s41416-020-0937-0.
C. Hou et al., “Predicting Breast Cancer in Chinese Women Using Machine Learning Techniques: Algorithm Development,” JMIR Medical Informatics, vol. 8, no. 6, p. e17364, Jun. 2020, DOI: 10.2196/17364.
A. Baltres et al., “Prediction of Oncotype DX recurrence score using deep multi-layer perceptrons in estrogen receptor-positive, HER2-negative breast cancer,” Breast Cancer, vol. 27, no. 5, pp. 1007–1016, May 2020, DOI: 10.1007/s12282-020-01100-4.
G. Yuan, Y. Liu, W. Huang, and B. Hu, “Differentiating Grade in Breast Invasive Ductal Carcinoma Using Texture Analysis of MRI,” Computational and Mathematical Methods in Medicine, vol. 2020, pp. 1–14, Apr. 2020, DOI: 10.1155/2020/6913418.
L. Yang et al., “Prediction model of the response to neoadjuvant chemotherapy in breast cancers by a Naive Bayes algorithm,” Computer Methods and Programs in Biomedicine, vol. 192, p. 105458, Aug. 2020, DOI: 10.1016/j.cmpb.2020.105458.
C. Nicolò et al., “Machine Learning and Mechanistic Modeling for Prediction of Metastatic Relapse in Early-Stage Breast Cancer,” JCO Clinical Cancer Informatics, vol. 1, no. 4, pp. 259–274, Sep. 2020, DOI: 10.1200/cci.19.00133.
A. Fanizzi et al., “A machine learning approach on multiscale texture analysis for breast microcalcification diagnosis,” BMC Bioinformatics, vol. 21, no. S2, pp. 1–11, Mar. 2020, DOI: 10.1186/s12859-020-3358-4.
C. Boeri et al., “Machine Learning techniques in breast cancer prognosis prediction: A primary evaluation,” Cancer Medicine, vol. 1, no. 1, pp. 3234–3243, Mar. 2020, DOI: 10.1002/cam4.2811.
V. Madekivi, P. Boström, A. Karlsson, R. Aaltonen, and E. Salminen, “Can a machine-learning model improve the prediction of nodal stage after a positive sentinel lymph node biopsy in breast cancer?,” Acta Oncologica, vol. 59, no. 6, pp. 689–695, Mar. 2020, DOI: 10.1080/0284186x.2020.1736332.
R. Castaldo, K. Pane, E. Nicolai, M. Salvatore, and M. Franzese, “The Impact of Normalization Approaches to Automatically Detect Radiogenomic Phenotypes Characterizing Breast Cancer Receptors Status,” Cancers, vol. 12, no. 2, p. 518, Feb. 2020, DOI: 10.3390/cancers12020518.
Z. Salod and Y. Singh, “Comparison of the performance of machine learning algorithms in breast cancer screening and detection: A protocol,” Journal of Public Health Research, vol. 8, no. 3, pp. 112–118, Dec. 2019, DOI: 10.4081/jphr.2019.1677.
Y. Xu, L. Ju, J. Tong, C. Zhou, and J. Yang, “Supervised Machine Learning Predictive Analytics For Triple-Negative Breast Cancer Death Outcomes,” OncoTargets and therapy, vol. 12, no. 1, pp. 9059–9067, Nov. 2019, DOI: 10.2147/OTT.S223603.
E. K. Park et al., “Machine Learning Approaches to Radiogenomics of Breast Cancer using Low-Dose Perfusion Computed Tomography: Predicting Prognostic Biomarkers and Molecular Subtypes,” Scientific Reports, vol. 9, no. 1, pp. 1–11, Nov. 2019, DOI: 10.1038/s41598-019-54371-z.
N. L. Eun et al., “Texture Analysis with 3.0-T MRI for Association of Response to Neoadjuvant Chemotherapy in Breast Cancer,” Radiology, vol. 294, no. 1, pp. 31–41, Jan. 2020, DOI: 10.1148/radiol.2019182718.
L.Q. Zhou et al., “Lymph Node Metastasis Prediction from Primary Breast Cancer US Images Using Deep Learning,” Radiology, vol. 294, no. 1, pp. 19–28, Jan. 2020, DOI: 10.1148/radiol.2019190372.
M. Hsieh, L. Sun, C. Lin, M. Hsieh, C. Hsu, and C. Kao, “The Performance of Different Artificial Intelligence Models in Predicting Breast Cancer among Individuals Having Type 2 Diabetes Mellitus,” Cancers, vol. 11, no. 11, p. 1751, Nov. 2019, DOI: 10.3390/cancers11111751.
A. Bomane, A. Gonçalves, and P. J. Ballester, “Paclitaxel Response Can Be Predicted With Interpretable Multi-Variate Classifiers Exploiting DNA-Methylation and miRNA Data,” Frontiers in Genetics, vol. 10, p. 1041, Oct. 2019, DOI: 10.3389/fgene.2019.01041.
J. Liu et al., “Radiomics Analysis of Dynamic Contrast-Enhanced Magnetic Resonance Imaging for the Prediction of Sentinel Lymph Node Metastasis in Breast Cancer,” Frontiers in Oncology, vol. 9, pp. 1–8, Sep. 2019, DOI: 10.3389/fonc.2019.00980.
H. Chereda, A. Bleckmann, F. Kramer, A. Leha, and T. Beissbarth, “Utilizing Molecular Network Information via Graph Convolutional Neural Networks to Predict Metastatic Event in Breast Cancer,” Studies in Health Technology and Informatics, vol. 267, pp. 181–186, Sep. 2019, DOI: 10.3233/SHTI190824.
R. Kleinlein and D. Riaño, “Persistence of data-driven knowledge to predict breast cancer survival,” International Journal of Medical Informatics, vol. 129, pp. 303–311, Sep. 2019, DOI: 10.1016/j.ijmedinf.2019.06.018.
S. Bhattarai et al., “Machine learning-based prediction of breast cancer growth rate in vivo,” British Journal of Cancer, vol. 121, no. 6, pp. 497–504, Aug. 2019, DOI: 10.1038/s41416-019-0539-x.
H. Shimizu and K. I. Nakayama, “A 23 gene–based molecular prognostic score precisely predicts overall survival of breast cancer patients,” EBioMedicine, vol. 46, pp. 150–159, Aug. 2019, DOI: 10.1016/j.ebiom.2019.07.046.
G. Shamai, Y. Binenbaum, R. Slossberg, I. Duek, Z. Gil, and R. Kimmel, “Artificial Intelligence Algorithms to Assess Hormonal Status From Tissue Microarrays in Patients With Breast Cancer,” JAMA Network Open, vol. 2, no. 7, p. e197700, Jul. 2019, DOI: 10.1001/jamanetworkopen.2019.7700.
F. J. Pérez-Benito et al., “Global parenchymal texture features based on histograms of oriented gradients improve cancer development risk estimation from healthy breasts,” Computer Methods and Programs in Biomedicine, vol. 177, pp. 123–132, Aug. 2019, DOI: 10.1016/j.cmpb.2019.05.022.
T. Murata et al., “Salivary metabolomics with alternative decision tree-based machine learning methods for breast cancer discrimination,” Breast Cancer Research and Treatment, vol. 177, no. 3, pp. 591–601, Jul. 2019, DOI: 10.1007/s10549-019-05330-9.
C. Ming, V. Viassolo, N. Probst-Hensch, P. O. Chappuis, I. D. Dinov, and M. C. Katapodi, “Machine learning techniques for personalized breast cancer risk prediction: comparison with the BCRAT and BOADICEA models,” Breast cancer research: BCR, vol. 21, no. 1, p. 75, Jun. 2019, DOI: 10.1186/s13058-019-1158-4.
T. He et al., “A Deep Learning–Based Decision Support Tool for Precision Risk Assessment of Breast Cancer,” JCO Clinical Cancer Informatics, no. 3, pp. 1–12, Dec. 2019, DOI: 10.1200/cci.18.00121.
R. Turkki et al., “Breast cancer outcome prediction with tumour tissue images and machine learning,” Breast Cancer Research and Treatment, vol. 177, pp. 41–52, May 2019, DOI: 10.1007/s10549-019-05281-1.
M. Wu et al., “Prediction of molecular subtypes of breast cancer using BI-RADS features based on a ‘white box’ machine learning approach in a multi-modal imaging setting,” European Journal of Radiology, vol. 114, pp. 175–184, May 2019, DOI: 10.1016/j.ejrad.2019.03.015.
A. A. Tabl, A. Alkhateeb, W. ElMaraghy, L. Rueda, and A. Ngom, “A Machine Learning Approach for Identifying Gene Biomarkers Guiding the Treatment of Breast Cancer,” Frontiers in Genetics, vol. 10, pp. 1–13, Mar. 2019, DOI: 10.3389/fgene.2019.00256.
J. You, R. D. McLeod, and P. Hu, “Predicting drug-target interaction network using deep learning model,” Computational Biology and Chemistry, vol. 80, pp. 90–101, Jun. 2019, DOI: 10.1016/j.compbiolchem.2019.03.016.
M. D. Ganggayah, N. A. Taib, Y. C. Har, P. Lio, and S. K. Dhillon, “Predicting factors for survival of breast cancer patients using machine learning techniques,” BMC Medical Informatics and Decision Making, vol. 19, no. 1, pp. 1–17, Mar. 2019, DOI: 10.1186/s12911-019-0801-4.
T. Turki and J. T. L. Wang, “Clinical intelligence: New machine learning techniques for predicting clinical drug response,” Computers in Biology and Medicine, vol. 107, pp. 302–322, Apr. 2019, DOI: 10.1016/j.compbiomed.2018.12.017.
Z. Zeng et al., “Using natural language processing and machine learning to identify breast cancer local recurrence,” BMC Bioinformatics, vol. 19, no. S17, pp. 65–74, Dec. 2018, DOI: 10.1186/s12859-018-2466-x.
C. Nickson et al., “Prospective validation of the NCI Breast Cancer Risk Assessment Tool (Gail Model) on 40,000 Australian women,” Breast Cancer Research, vol. 20, no. 1, pp. 1–12, Dec. 2018, DOI: 10.1186/s13058-018-1084-x.
N. Mao et al., “Added Value of Radiomics on Mammography for Breast Cancer Diagnosis: A Feasibility Study,” Journal of the American College of Radiology, vol. 16, no. 4, pp. 485–491, Apr. 2019, DOI: 10.1016/j.jacr.2018.09.041.
D. S. Lituiev et al., “Automatic Labeling of Special Diagnostic Mammography Views from Images and DICOM Headers,” Journal of Digital Imaging, vol. 32, no. 2, pp. 228–233, Nov. 2018, DOI: 10.1007/s10278-018-0154-z.
M. Zhao, Y. Tang, H. Kim, and K. Hasegawa, “Machine Learning With K-Means Dimensional Reduction for Predicting Survival Outcomes in Patients With Breast Cancer,” Cancer Informatics, vol. 17, no. 1, p. 117693511881021, Jan. 2018, DOI: 10.1177/1176935118810215.
Z. Zhang et al., “Morphology-based prediction of cancer cell migration using an artificial neural network and a random decision forest,” Integrative biology : quantitative biosciences from nano to macro, vol. 10, no. 12, pp. 758–767, 2018, DOI: 10.1039/c8ib00106e.
A. Tahmassebi et al., “Impact of Machine Learning With Multiparametric Magnetic Resonance Imaging of the Breast for Early Prediction of Response to Neoadjuvant Chemotherapy and Survival Outcomes in Breast Cancer Patients,” Investigative Radiology, vol. 54, no. 2, pp. 110–117, Feb. 2019, DOI: 10.1097/rli.0000000000000518.
I. Kim et al., “A predictive model for high/low risk group according to oncotype DX recurrence score using machine learning,” European Journal of Surgical Oncology, vol. 45, no. 2, pp. 134–140, Feb. 2019, DOI: 10.1016/j.ejso.2018.09.011.
E. H. Cain, A. Saha, M. R. Harowicz, J. R. Marks, P. K. Marcom, and M. A. Mazurowski, “Multivariate machine learning models for prediction of pathologic response to neoadjuvant therapy in breast cancer using MRI features: a study using an independent validation set,” Breast Cancer Research and Treatment, vol. 173, no. 2, pp. 455–463, Oct. 2018, DOI: 10.1007/s10549-018-4990-9.
M. Sherafatian, “Tree-based machine learning algorithms identified minimal set of miRNA biomarkers for breast cancer diagnosis and molecular subtyping,” Gene, vol. 677, no. 1, pp. 111–118, Nov. 2018, DOI: 10.1016/j.gene.2018.07.057.
W. Ma, Y. Ji, L. Qi, X. Guo, X. Jian, and P. Liu, “Breast cancer Ki67 expression prediction by DCE-MRI radiomics features,” Clinical Radiology, vol. 73, no. 10, pp. 909.e1–909.e5, Oct. 2018, DOI: 10.1016/j.crad.2018.05.027.
J. Lötsch et al., “Machine-learning-derived classifier predicts absence of persistent pain after breast cancer surgery with high accuracy,” Breast Cancer Research and Treatment, vol. 171, no. 2, pp. 399–411, Jun. 2018, DOI: 10.1007/s10549-018-4841-8.
W. Ma et al., “Breast Cancer Molecular Subtype Prediction by Mammographic Radiomic Features,” Academic Radiology, vol. 26, no. 2, pp. 196–201, Feb. 2019, DOI: 10.1016/j.acra.2018.01.023.
N. Shukla, M. Hagenbuchner, K. T. Win, and J. Yang, “Breast cancer data analysis for survivability studies and prediction,” Computer Methods and Programs in Biomedicine, vol. 155, no. 1, pp. 199–208, Mar. 2018, DOI: 10.1016/j.cmpb.2017.12.011.
H. Jiang, W.-K. Ching, W.-S. Cheung, W. Hou, and H. Yin, “Hadamard Kernel SVM with applications for breast cancer outcome predictions,” BMC Systems Biology, vol. 11, no. S7, pp. 163–174, Dec. 2017, DOI: 10.1186/s12918-017-0514-1.
M. Patrício et al., “Using Resistin, glucose, age and BMI to predict the presence of breast cancer,” BMC Cancer, vol. 18, no. 1, pp. 1–8, Jan. 2018, DOI: 10.1186/s12885-017-3877-1.
M. Heidari et al., “Prediction of breast cancer risk using a machine learning approach embedded with a locality preserving projection algorithm,” Physics in Medicine & Biology, vol. 63, no. 3, p. 035020, Jan. 2018, DOI: 10.1088/1361-6560/aaa1ca.
F. M. Alakwaa, K. Chaudhary, and L. X. Garmire, “Deep Learning Accurately Predicts Estrogen Receptor Status in Breast Cancer Metabolomics Data,” Journal of Proteome Research, vol. 17, no. 1, pp. 337–347, Nov. 2017, DOI: 10.1021/acs.jproteome.7b00595.
I. Vidić et al., “Support vector machine for breast cancer classification using diffusion-weighted MRI histogram features: Preliminary study,” Journal of Magnetic Resonance Imaging, vol. 47, no. 5, pp. 1205–1216, Oct. 2017, DOI: 10.1002/jmri.25873.
A. Chan and J. A. Tuszynski, “Automatic prediction of tumour malignancy in breast cancer with fractal dimension,” Royal Society Open Science, vol. 3, no. 12, p. 160558, Dec. 2016, DOI: 10.1098/rsos.160558.
M. Huang, C. Chen, W. Lin, S. Ke, and C. Tsai, “SVM and SVM Ensembles in Breast Cancer Prediction,” PloS one, vol. 12, no. 1, p. e0161501, 2017, DOI: 10.1371/journal.pone.0161501.
F. P. Y. Lin, A. Pokorny, C. Teng, R. Dear, and R. J. Epstein, “Computational prediction of multidisciplinary team decision-making for adjuvant breast cancer drug therapies: a machine learning approach,” BMC Cancer, vol. 16, no. 1, pp. 1–10, Dec. 2016, DOI: 10.1186/s12885-016-2972-z.
S. Vural, X. Wang, and C. Guda, “Classification of breast cancer patients using somatic mutation profiles and machine learning approaches,” BMC Systems Biology, vol. 10, no. S3, pp. 264–276, Aug. 2016, DOI: 10.1186/s12918-016-0306-z.
W. Kim, K. S. Kim, and R. W. Park, “Nomogram of Naive Bayesian Model for Recurrence Prediction of Breast Cancer,” Healthcare Informatics Research, vol. 22, no. 2, pp. 89–94, Apr. 2016, DOI: 10.4258/hir.2016.22.2.89.
M. Montazeri, M. Montazeri, M. Montazeri, and A. Beigzadeh, “Machine learning models in breast cancer survival prediction,” Technology and Health Care, vol. 24, no. 1, pp. 31–42, Jan. 2016, DOI: 10.3233/thc-151071.
S. N. Dorman et al., “Genomic signatures for paclitaxel and gemcitabine resistance in breast cancer derived by machine learning,” Molecular Oncology, vol. 10, no. 1, pp. 85–100, Aug. 2015, DOI: 10.1016/j.molonc.2015.07.006.