Senescence Reversion in Plant Images Using Perception and Unpaired Data
Keywords:
Image-to-image translation, Unpaired data, CycleGAN, Herbaria, Senescence reversionAbstract
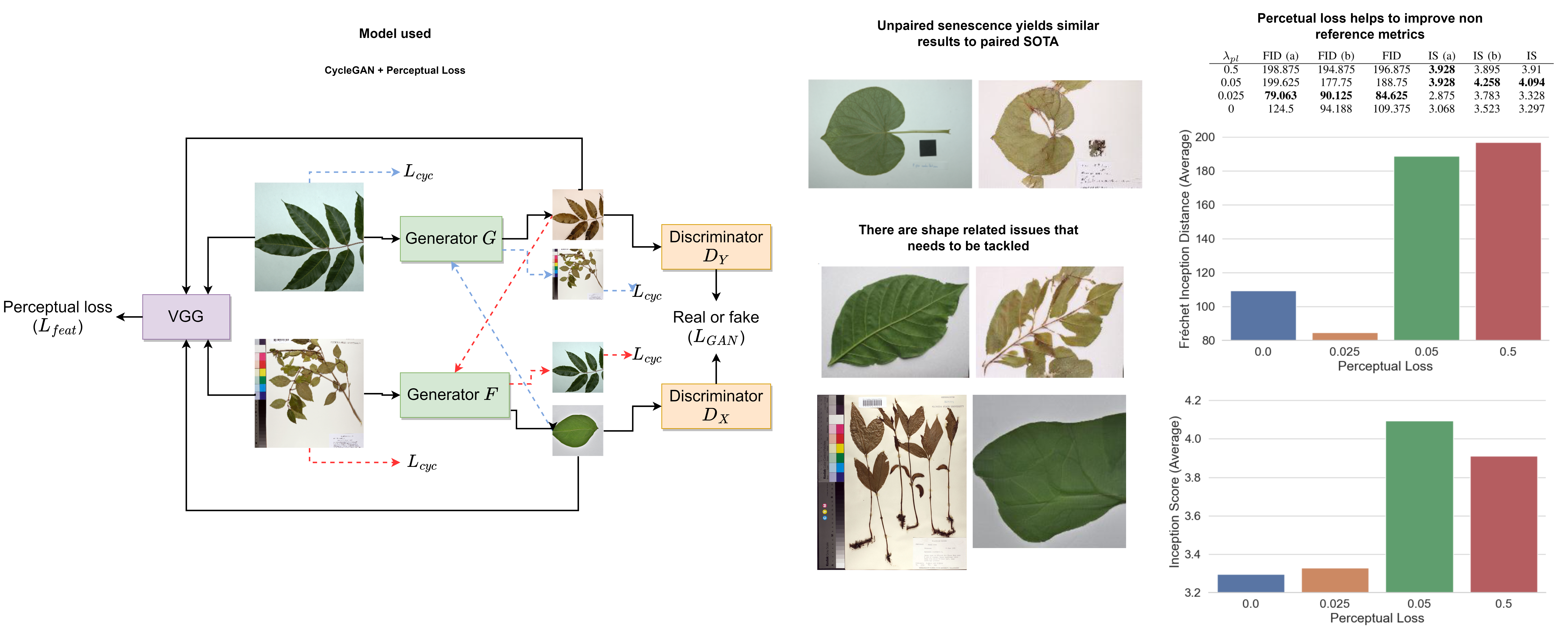
Recent work on using herbarium images for automatic plant identification, and in particular to do domain adaptation to field images, has been promising. A potential way to address such domain adaptation problem is generative: Hallucinate how a herbarium image would have looked like when it was in the field, and use such synthetic, fresh and green image for plant identification purposes. Such generative task, called senescence reversion, has been poorly explored. To our knowledge, it has been studied only in terms of paired data, meaning, with pairs of dry and fresh images of the same specimen. Such paired data is hard to produce, curate and find. In this work we explore herbarium senescence reversion via unpaired data. The lack of pairs at specimen level presents its own challenges, as capturing the intricacies of each species depends on images of different specimens from different domains. We explore learning a mapping from a herbarium image to leaf plant images and vice-versa, aligning two models, one for each herbarium and leaf domain. We experiment against the state-of-the-art paired baseline Pix2Pix which yields a SSIM of 0.8986, compared to our unpaired approach that yields 0.8865, showing very similar results on reconstruction metrics regardless of our approach not having the luxury of pairs by specimen. Additionally, we apply perceptual loss in order to improve the natural look of the synthetic images. The balance of how much perception is good to avoid reconstruction problems is also studied. Lastly, using a new unpaired dataset built by ourselves, our results show that using a low lambda value from 0.025 to 0.05 for perceptual loss, helps getting lower Fráchet Inception Distances and higher Inception Scores. To our knowledge, no other work has focused on reverting senescence of herbarium sheet images based on unpaired data.
Downloads
References
M. Yerman, “Herbarium Explorations,” American Gardener, 2017.
J. Carranza-Rojas, H. Goeau, P. Bonnet, E. MataMontero, and A. Joly, “Going deeper in the automated
identification of Herbarium specimens,” BMC Evolutionary Biology, vol. 17, no. 1, p. 181, Aug. 11, 2017,
ISSN: 1471-2148.
H. Goeau, P. Bonnet, and A. Joly, “Overview of LifeCLEF Plant Identification task 2020,” p. 15, 2020.
M.-Y. Liu, T. Breuel, and J. Kautz, “Unsupervised
Image-to-Image Translation Networks,” Jul. 22, 2018.
arXiv: 1703.00848 [cs].
Y. S. Chen, Y. C. Wang, M. H. Kao, and Y. Y. Chuang,
“Deep Photo Enhancer: Unpaired Learning for Image
Enhancement from Photographs with GANs,” Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, pp. 6306–6314,
, ISSN: 9781538664209.
A. Affouard, H. Goeau, P. Bonnet, J.-C. Lombardo, and ¨
A. Joly, “Pl@ntNet app in the era of deep learning,”
p. 7, 2017.
B. R. Hussein, O. A. Malik, W.-H. Ong, and J. W. F.
Slik, “Reconstruction of damaged herbarium leaves
using deep learning techniques for improving classification accuracy,” Ecological Informatics, vol. 61,
pp. 101 243–101 243, Feb. 2021.
J. Villacis-Llobet, M. Lucio-Troya, M. Calvo-Navarro,
S. Calderon-Ramirez, and E. Mata-Montero, “A First
Glance into Reversing Senescence on Herbarium Sample Images Through Conditional Generative Adversarial Networks,” in Communications in Computer and
Information Science, vol. 1087 CCIS, Springer, 2020,
pp. 438–447, ISBN: 978-3-030-41004-9.
J. Y. Zhu, T. Park, P. Isola, and A. A. Efros,
“Unpaired Image-to-Image Translation Using CycleConsistent Adversarial Networks,” 9781538610329,
, pp. 2242–2251.
H. Talebi and P. Milanfar, “NIMA: Neural Image Assessment,” IEEE Transactions on Image Processing,
vol. 27, no. 8, pp. 3998–4011, 2018.
Y. Blau and T. Michaeli, “The Perception-Distortion
Tradeoff,” 2018.
P. Isola, J. Y. Zhu, T. Zhou, and A. A. Efros, “Imageto-image translation with conditional adversarial networks,” 9781538604571, 2017, pp. 5967–5976.
I. J. Goodfellow, J. Pouget-Abadie, M. Mirza, et al.,
“Generative Adversarial Networks,” Jun. 2014.
Z. Wang, A. Bovik, H. Sheikh, and E. Simoncelli,
“Image Quality Assessment: From Error Visibility to
Structural Similarity,” IEEE Transactions on Image Processing, vol. 13, no. 4, pp. 600–612, Apr. 2004, ISSN:
-7149.
O. Ronneberger, P. Fischer, and T. Brox, “U-Net: Convolutional Networks for Biomedical Image Segmentation,” May 18, 2015. arXiv: 1505.04597 [cs].
K. Simonyan and A. Zisserman, “Very Deep Convolutional Networks for Large-Scale Image Recognition,”
Apr. 10, 2015. arXiv: 1409.1556 [cs].
M. Ounsworth. “Anticipatory movement planning for
quadrotor visual servoeing.” in collab. with G. L. Dudek
(Internal/Supervisor). (), [Online]. Available: https : / /
escholarship . mcgill . ca / concern / theses / zc77ss96z
(visited on 05/26/2021).
X. Mao, Q. Li, H. Xie, R. Y. K. Lau, Z. Wang, and
S. P. Smolley, “Least Squares Generative Adversarial
Networks,” Apr. 5, 2017. arXiv: 1611.04076 [cs].
X. Huang, M.-Y. Liu, S. Belongie, and J. Kautz,
“Multimodal Unsupervised Image-to-Image Translation,” Aug. 14, 2018. arXiv: 1804 . 04732 [cs,
stat].
M. Amodio and S. Krishnaswamy, “TraVeLGAN:
Image-to-image Translation by Transformation Vector
Learning,” Feb. 25, 2019. arXiv: 1902.09631 [cs].
T. Wang and Y. Lin, “CycleGAN with Better Cycles,”
p. 8,
T. Park, A. A. Efros, R. Zhang, and J.-Y. Zhu, “Contrastive Learning for Unpaired Image-to-Image Translation,” Aug. 20, 2020. arXiv: 2007.15651 [cs].
T. Chen, S. Kornblith, M. Norouzi, and G. Hinton, “A
Simple Framework for Contrastive Learning of Visual
Representations,” Jun. 30, 2020. arXiv: 2002 . 05709
[cs, stat].
J. Johnson, A. Alahi, and L. Fei-Fei, “Perceptual
losses for real-time style transfer and super-resolution,”
, 2016, pp. 694–711.
R. Zhang, P. Isola, A. A. Efros, E. Shechtman, and O.
Wang, “The Unreasonable Effectiveness of Deep Features as a Perceptual Metric,” 9781538664209, 2018,
pp. 586–595.
J. Deng, W. Dong, R. Socher, L.-J. Li, K. Li, and
L. Fei-Fei, “ImageNet: A large-scale hierarchical image database,” in 2009 IEEE Conference on Computer
Vision and Pattern Recognition, Jun. 2009, pp. 248–255.