Simplifying VGG-16 for Plant Species Identification
Keywords:
Convolutional Neural Network, Deep Learning, Fine-grained Classification, Plant Species Identification, VGG-16Abstract
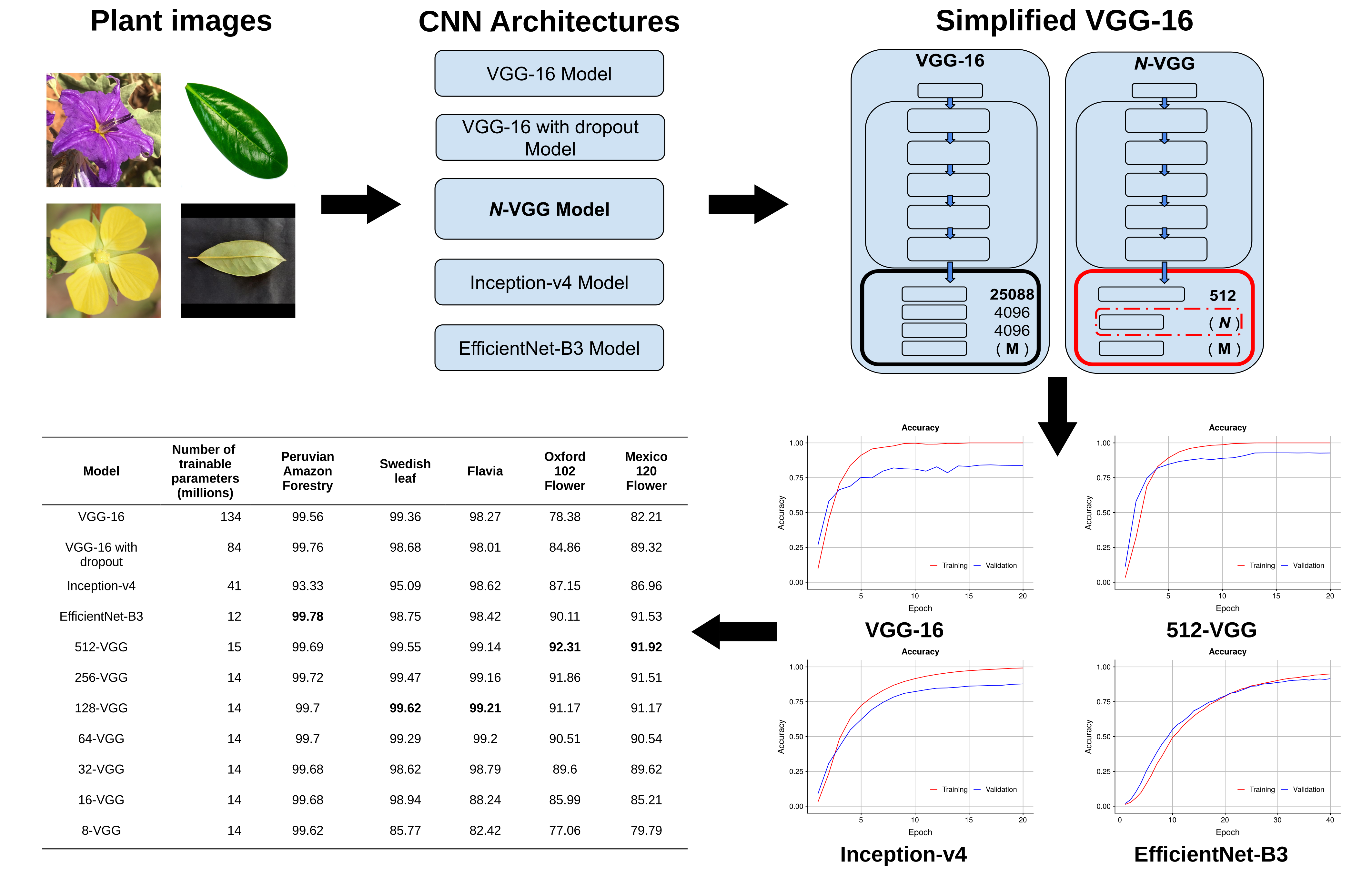
Plant species identification represents an extraordinary challenge for machine learning due to visual interspecies similarities and large intraspecies variations. Furthermore, research literature reports that plant species identification usually lacks sufficiently large datasets for training classification models. In this paper, we address this problem with a model that simplifies the VGG-16 architecture, the N-VGG model. The idea behind N-VGG is to reduce experimentally observed overfitting on VGG-16 by using as few trainable parameters as possible. To do this, we substitute the flattening layer on the VGG architecture with a global average pooling layer. This reduces the size of the feature vector. In addition, we eliminate one of the two fully-connected layers and use a new hyper-parameter, N, to indicate the number of nodes on the remaining layer. To show the robustness of the N-VGG model, we conducted extensive experimentation. We trained N-VGG on five datasets for plant species identification. Four of these datasets are publicly available and have been widely used as benchmarks for plant identification models. For all datasets, we compare the accuracy of N-VGG to that of the VGG-16, Inception-v4, and EfficienNet-B3 models. The experimental results show that the N-VGG model achieved the best classification performance for all but one datasets, whereas all the models showed a remarkable performance for the remaining dataset. This evidence supports our initial idea that, for plant species classification, some accuracy might be lost due to overfitting and that having fewer trainable parameters helps in producing a more robust model.
Downloads
References
Y. LeCun, Y. Bengio, and G. Hinton, “Deep Learning.,” Nature, vol. 521, no. 7553, pp. 436–444, 2015.
I. Goodfellow, Y. Bengio, and A. Courville, Deep Learning. MIT Press, 2016.
A. Khan, A. Sohail, U. Zahoora, and A. S. Qureshi, “A Survey of the Recent Architectures of Deep Convolutional Neural Networks.,” Artificial Intelligence Review, vol. 53, no. 8, pp. 5455–5516, 2020.
M. Mehdipour-Ghazi, B. A. Yanikoglu, and E. Aptoula, “Plant Identification Using Deep Neural Networks Via Optimization of Transfer Learning Parameters.,” Neurocomputing, vol. 235, pp. 228–235, 2017.
K. R. Aravind, P. Raja, R. Ashiwin, and K. V. Mukesh, “Disease Classification in Solanum Melongena Using Deep Learning.,” Spanish Journal of Agricultural Research, vol. 17, no. 3, 2019. https://doi.org/10.5424/sjar/2019173-14762.
Y. Li, J. Nie, and X. Chao, “Do we Really Need Deep CNN for Plant Diseases Identification?,” Computers and Electronics in Agriculture, vol. 178, 2020. https://doi.org/10.1016/j.compag.2020.105803.
S. H. Lee, H. Goëau, P. Bonnet, and A. Joly, “New Perspectives on Plant Disease Characterization Based on Deep Learning.,” Computers and Electronics in Agriculture, vol. 170, 2020. https://doi.org/10.1016/j.compag.2020.105220.
J. M. Johnson and T. M. Khoshgoftaar, “Survey on Deep Learning with Class Imbalance.,” Journal of Big Data, vol. 6, no. 1, pp. 27–27, 2019.
A.-X. Li, K.-X. Zhang, and L.-W. Wang, “Zero-Shot Fine-Grained Classification by Deep Feature Learning with Semantics.,” International Journal of Automation and Computing, vol. 16, no. 5, pp. 563–574, 2019.
A. Kaya, A. S. Keceli, C. Catal, H. Y. Yalic, H. Temucin, and B. Tekinerdogan, “Analysis of Transfer Learning for Deep Neural Network Based Plant Classification Models.,” Computers and Electronics in Agriculture, vol. 158, pp. 20–29, 2019.
F. Chollet, Deep learning with Python. Manning Publications, 2017.
B. Zhou, A. Khosla, À. Lapedriza, A. Oliva, and A. Torralba, “Learning Deep Features for Discriminative Localization.,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, June 27-30, pp. 2921–2929, IEEE Computer Society, 2016.
P. Barré, B. C. Stöver, K. F. Müller, and V. Steinhage, “Leafsnap: A Computer Vision System for Automatic Plant Species Identification.,” Ecological Informatics, vol. 40, pp. 50–56, 2017.
Y. Lu, S. Yi, N. Zeng, Y. Liu, and Y. Zhang, “Identification of Rice Diseases using Deep Convolutional Neural Networks.,” Neurocomputing, vol. 267, pp. 378–384, 2017.
E. C. Too, L. Yujian, S. Njuki, and L. Yingchun, “A Comparative Study of Fine-tuning Deep Learning Models for Plant Disease Identification.,” Computers and Electronics in Agriculture, vol. 161, pp. 272–279, 2019.
J. Deng, W. Dong, R. Socher, L. Li, K. Li, and L. Fei-Fei, “Imagenet: A Large-scale Hierarchical Image Database.,” in Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Miami, Florida, USA, June 20-25 , pp. 248–255, IEEE Computer Society, 2009.
M. Everingham, L. Van Gool, C. K. Williams, J. Winn, and A. Zisserman, “The Pascal Visual Object Classes (VOC) Challenge.,” International Journal of Computer Vision, vol. 88, no. 2, pp. 303–338, 2010.
T. Lin, M. Maire, S. J. Belongie, J. Hays, P. Perona, D. Ramanan, P. Dollár, and C. L. Zitnick, “Microsoft COCO: Common Objects in Context.,” in Proceedings of the European Conference on Computer Vision, Zurich, Switzerland, September 6-12 (D. J. Fleet, T. Pajdla, B. Schiele, and T. Tuytelaars, eds.), vol. 8693 of Lecture Notes in Computer Science, pp. 740–755, Springer, 2014.
A. Joly, H. Goëau, H. Glotin, C. Spampinato, P. Bonnet, W. Vellinga, R. Planqué, A. Rauber, S. Palazzo, B. Fisher, and H. Müller, “LifeCLEF 2015: Multimedia Life Species Identification Challenges.,” in Proceedings of the International Conference of the CLEF Association, Toulouse, France, September 8-11 (J. Mothe, J. Savoy, J. Kamps, K. Pinel-Sauvagnat, G. J. F. Jones, E. SanJuan, L. Cappellato, and N. Ferro, eds.), vol. 9283 of Lecture Notes in Computer Science, pp. 462–483, Springer, 2015.
H. Goëau, P. Bonnet, and A. Joly, “Plant Identification in an Open-World (lifeCLEF 2016).,” in Proceedings of the Conference and Labs of the Evaluation forum, Évora, Portugal, September 5-8 (K. Balog, L. Cappellato, N. Ferro, and C. Macdonald, eds.), vol. 1609 of CEUR Workshop Proceedings, pp. 428–439, CEUR-WS, 2016.
H. Goëau, P. Bonnet, and A. Joly, “Plant Identification Based on Noisy Web Data: the Amazing Performance of Deep Learning (LifeCLEF 2017).,” in Proceedings of the Conference and Labs of the Evaluation Forum, Dublin, Ireland, September 11-14 (L. Cappellato, N. Ferro, L. Goeuriot, and T. Mandl, eds.), vol. 1866 of CEUR Workshop Proceedings, CEUR-WS, 2017.
H. Goëau, P. Bonnet, and A. Joly, “Overview of ExpertLifeCLEF 2018: How far Automated Identification Systems are from the Best Experts?,” in Proceedings of the Conference and Labs of the Evaluation Forum, Avignon, France, September 10-14 (L. Cappellato, N. Ferro, J. Nie, and L. Soulier, eds.), vol. 2125 of CEUR Workshop Proceedings, CEUR-WS, 2018.
A. Krizhevsky, I. Sutskever, and G. E. Hinton, “Imagenet Classification with Deep Convolutional Neural Networks.,” Communications of the ACM, vol. 60, no. 6, pp. 84–90, 2017.
K. Simonyan and A. Zisserman, “Very Deep Convolutional Networks for Large-Scale Image Recognition.,” in Proceedings of the International Conference on Learning Representations, San Diego, CA, USA, May 7-9 (Y. Bengio and Y. LeCun, eds.), 2015.
C. Szegedy, W. Liu, Y. Jia, P. Sermanet, S. E. Reed, D. Anguelov, D. Erhan, V. Vanhoucke, and A. Rabinovich, “Going Deeper with Convolutions.,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, June 7-12, pp. 1–9, IEEE Computer Society, 2015.
K. He, X. Zhang, S. Ren, and J. Sun, “Deep Residual Learning for Image Recognition.,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, June 27-30, pp. 770–778, IEEE Computer Society, 2016.
C. Szegedy, S. Ioffe, V. Vanhoucke, and A. A. Alemi, “Inception-v4, Inception-ResNet and the Impact of Residual Connections on Learning.,” in Proceedings of the AAAI Conference on Artificial Intelligence, San Francisco, California, USA, February 4, Association for the Advancement of Artificial Intelligence, p. 4278–4284, AAAI Press, 2017.
F. Chollet, “Xception: Deep Learning with Depthwise Separable Convolutions.,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, July 21-26, pp. 1800–1807, IEEE Computer Society, 2017.
M. Sandler, A. G. Howard, M. Zhu, A. Zhmoginov, and L. Chen, “MobileNetV2: Inverted Residuals and Linear Bottlenecks,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, June 18-22, pp. 4510–4520, Computer Vision Foundation / IEEE Computer Society, 2018.
M. Sulc, L. Picek, and J. Matas, “Plant Recognition by Inception Networks with Test-time Class Prior Estimation.,” in Proceedings of the Conference and Labs of the Evaluation Forum, Avignon, France, September 10-14 (L. Cappellato, N. Ferro, J. Nie, and L. Soulier, eds.), vol. 2125 of CEUR Workshop Proceedings, CEUR-WS, 2018.
J. W. Lee and Y. C. Yoon, “Fine-Grained Plant Identification using Wide and Deep Learning Model.,” in Proceedings of the International Conference on Platform Technology and Service, Jeju, Korea (South), January 28-30, pp. 1–5, IEEE Computer Society, 2019.
S. I. Saedi and H. Khosravi, “A Deep Neural Network Approach Towards Real-Time on-branch Fruit Recognition for Precision Horticulture.,” Expert Systems With Applications, vol. 159, 2020. https://doi.org/10.1016/j.eswa.2020.113594.
D. P. Hughes and M. Salathé, “An open Access Repository of Images on Plant Health to Enable the Development of Mobile Disease Diagnostics through Machine Learning and Crowdsourcing.,” Computing Research Repository, vol. abs/1511.08060, 2015.
G. Vizcarra, D. Bermejo, A. Mauricio, R. Z. Gomez, and E. Dianderas, “The Peruvian Amazon Forestry Dataset: A Leaf Image Classification Corpus.,” Ecological Informatics, vol. 62, 2021. https://doi.org/10.1016/j.ecoinf.2021.101268.
D. Bisen, “Deep Convolutional Neural Network Based Plant Species Recognition through Features of Leaf,” Multimedia Tools and Applications, vol. 80, no. 4, pp. 6443–6456, 2021.
O. Söderkvist, “Computer Vision Classification of Leaves from Swedish Trees,” Master’s thesis, Linkoping University, 2001.
L. A. D. Filho and R. T. Calumby, “An Experimental Assessment of Deep Convolutional Features for Plant Species Recognition.,” Ecological Informatics, vol. 65, 2021. https://doi.org/10.1016/j.ecoinf.2021.101411.
H. Goëau, A. Joly, P. Bonnet, V. Bakic, D. Barthélémy, N. Boujemaa, and J. Molino, “The Imageclef Plant Identification Task 2013.,” in Proceedings of the ACM International Workshop on Multimedia Analysis for Ecological Data, Barcelona, Spain, October 22 (C. Spampinato, V. Mezaris, and J. van Ossenbruggen, eds.), pp. 23–28, ACM, 2013.
Naturalista, “Comisión Nacional para el Conocimiento y Uso de la Biodiversidad.” http://www.naturalista.mx, 2021. Accessed: 2021-03-20.
S. G. Wu, F. S. Bao, E. Y. Xu, Y.-X. Wang, Y.-F. Chang, and Q.-L. Xiang, “A Leaf Recognition Algorithm for Plant Classification Using Probabilistic Neural Network.,” in Proceedings of the IEEE International Symposium on Signal Processing and Information Technology, Giza, Egypt, December 15-18, pp. 11–16, IEEE Computer Society, 2007.
M. Nilsback and A. Zisserman, “Automated Flower Classification over a Large Number of Classes.,” in Proceedings in the Indian Conference on Computer Vision, Graphics & Image Processing, ICVGIP 2008, Bhubaneswar, India, December 16-19, pp. 722–729, IEEE Computer Society, 2008.
M. Tan and Q. V. Le, “EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks,” in Proceedings of the 36th Interna tional Conference on Machine Learning, ICML, Long Beach, California, USA, June 9-15 (K. Chaudhuri and R. Salakhutdinov, eds.), vol. 97 of Proceedings of Machine Learning Research, pp. 6105–6114, PMLR, 2019.
N. Srivastava, G. E. Hinton, A. Krizhevsky, I. Sutskever, and R. Salakhutdinov, “Dropout: A Simple Way to Prevent Neural Networks from Overfitting,” J. Mach. Learn. Res., vol. 15, no. 1, pp. 1929–1958, 2014.