Brain Extraction in Multiple T1-weighted Magnetic Resonance Imaging slices using Digital Image Processing techniques
Keywords:
Image Processing, Skull Stripping, Brain Extraction, Image Segmentation, Medical ImagingAbstract
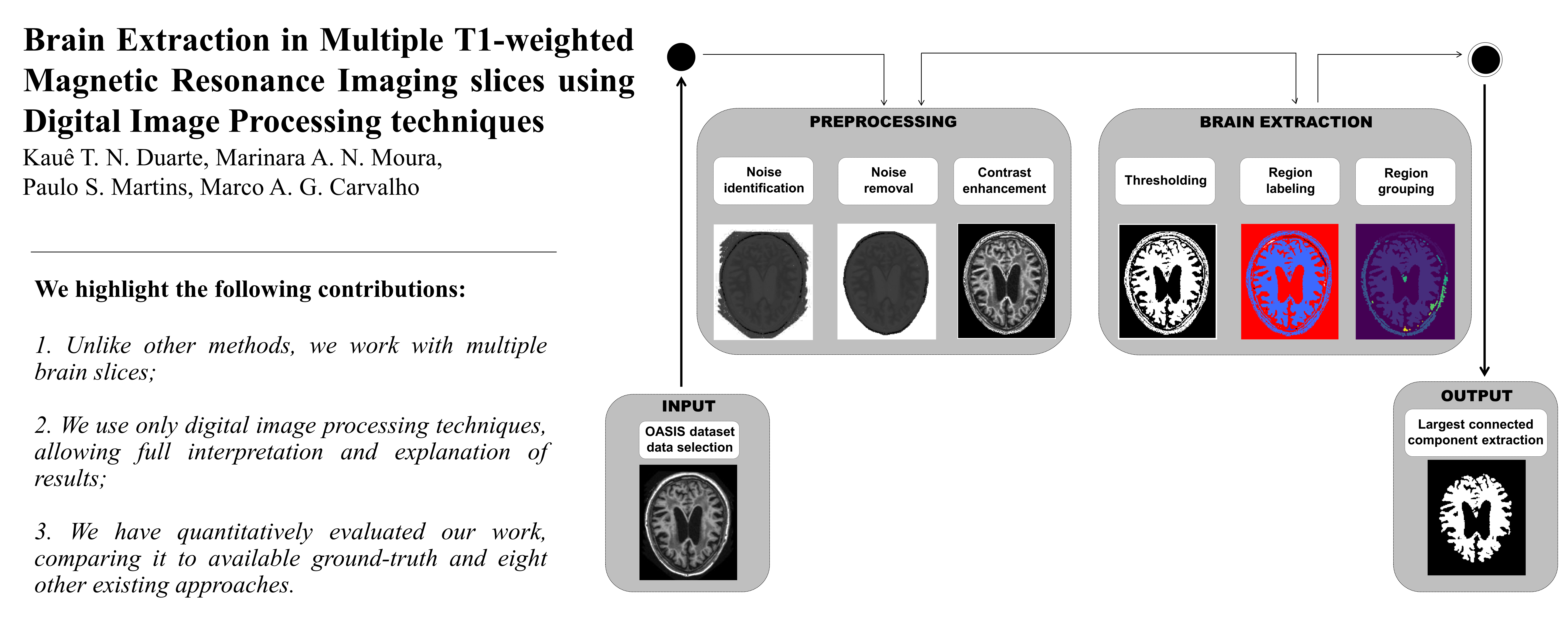
Brain Imaging has been source of several studies in the literature, mostly due to its importanceboth to predict and to analyze certain diseases or conditions. Extracting the brain from patient images for medical analysis can provide useful diagnostic and prognostic information.To this end, digital image processing algorithms have been applied to medical tasks with a focus on the identification of the brain. This work proposes a brain extraction framework based on three major steps: 1) Dataset and Image Selection; 2) Preprocessing; and 3) Largest Connected Component extraction. Our data are obtained from the OASIS dataset.The preprocessing step is applied in order to enhance contrast and eliminate possible noise from the T1-weighted MRI. Largest Connected Component extraction is performed by initially detecting the largest element in the image (i.e. the brain gray matter) and then by extracting it through mathematical morphology operators. The unsupervised framework extracts the brain in different axial slices without adjustments. The main contribution of this work is a method using only digital image processing for automatically identifying the brain from several different slices, which differs from the literature since is performed without parameter resetting. Five metrics were applied to evaluate our results: Specificity, Recall, Accuracy, F-Measure, and Precision. In our first experiment, two metrics resulted in more than 90% in efficiency (Specificity and Precision), two of them surpassed 80% (F-Measure and Accuracy), and Sensitivity exceeded 70%. Our second experiment compares our results with those produced by related works, having been ranked in the top positions of Sensitivity and Specificity.
Downloads
References
F. A. Azevedo, L. R. Carvalho, L. T. Grinberg, J. M. Farfel, R. E. Ferretti, R. E. Leite, W. J. Filho, R. Lent, and S. Herculano-Houzel, “Equal numbers of neuronal and nonneuronal cells make the human
brain an isometrically scaled-up primate brain,” vol. 513, pp. 532–541, Apr. 2009.
A. A. Ivanova, S. Srikant, Y. Sueoka, H. H. Kean, R. Dhamala, U.-M.O'Reilly, M. U. Bers, and E. Fedorenko, “Comprehension of computer code relies primarily on domain-general executive brain regions,” vol. 9, Dec. 2020.
A. S. Lundervold and A. Lundervold, “An overview of deep learning in medical imaging focusing on mri,” Zeitschrift für Medizinische Physik, vol. 29, no. 2, pp. 102–127, 2019. Special Issue: Deep Learning in Medical Physics.
T. Mohammad, “Skull removal in mr images using a modified artificial bee colony optimization algorithm,” Technology and Health Care, vol. 22, no. 5, p. 775–784, 2014.
J. Kleesiek, G. Urban, A. Hubert, D. Schwarz, K. Maier-Hein, M. Bendszus, and A. Biller, “Deep mri brain extraction: A 3d convolutional neural network for skull stripping,” NeuroImage, vol. 129, pp. 460 –
, 2016.
R. Roslan, N. Jamil, and R. Mahmud, “Skull stripping magnetic resonance images brain images : Region growing versus mathematical morphology,” 2011.
S. A. Sadananthan, W. Zheng, M. W. Chee, and V. Zagorodnov, “Skull stripping using graph cuts,” NeuroImage, vol. 49, no. 1, pp. 225 – 239, 2010.
A. Lakshmi, T. Arivoli, and R. Vinupriyadharshini, “Noise and skull removal of brain magnetic resonance image using curvelet transform and mathematical morphology,” in 2014 International Conference on Electronics and Communication Systems (ICECS), pp. 1–4, Feb 2014.
B. C. C. and L. V. L., “Morphology based enhancement and skull stripping of mri brain images,” in 2014 International Conference on Intelligent Computing Applications, pp. 254–257, March 2014.
K. Somasundaram and T. Kalaiselvi, “Automatic brain extraction methods for t1 magnetic resonance images using region labeling and morphological operations,” Computers in Biology and Medicine, vol. 41, no. 8, pp. 716 – 725, 2011.
A. S. Bhadauria, V. Bhateja, M. Nigam, and A. Arya, “Skull stripping of brain mri using mathematical morphology,” in Smart Intelligent Computing and Applications (S. C. Satapathy, V. Bhateja, J. R. Mohanty, and S. K. Udgata, eds.), (Singapore), pp. 775–780, Springer Singapore, 2020.
J. ´Swiebocka Wiek, “Skull stripping for mri images using morphological operators,” vol. 9842, pp. 172–182, 09 2016.
K. Srinivasan and N. Nanditha, “An intelligent skull stripping algorithm for mri image sequences using mathematical morphology.,” Biomedical Research (0970-938X), vol. 29, no. 16, 2018.
M. Laha, P. C. Tripathi, and S. Bag, “A skull stripping from brain mri using adaptive iterative thresholding and mathematical morphology,” in 2018 4th International Conference on Recent Advances in Information Technology (RAIT), pp. 1–6, 2018.
A. G. Balan, A. J. Traina, M. X. Ribeiro, P. M. Marques, and C. T. Jr., “Smart histogram analysis applied to the skull-stripping problem in t1-weighted mri,” Computers in Biology and Medicine, vol. 42, no. 5,
pp. 509 – 522, 2012.
A. Subudhi, J. Jena, and S. Sabut, “Extraction of brain from mri images by skull stripping using histogram partitioning with maximum entropy divergence,” in 2016 International Conference on Communication and Signal Processing (ICCSP), pp. 0931–0935, April 2016.
Z. Ullah, S.-H. Lee, and D. An, “Histogram equalization based enhancement and mr brain image skull stripping using mathematical morphology,” Authorea Preprints, 2020.
D. Genish and C. Balaji, “A novel skull stripping tool based on mean shift clustering and mathematical morphology,” 2021.
S. Roy and P. Maji, “An accurate and robust skull stripping method for 3-d magnetic resonance brain images,” Magnetic Resonance Imaging, vol. 54, pp. 46 – 57, 2018.
P. Kalavathi and V. B. S. Prasath, “Methods on skull stripping of MRI head scan images: a review,” vol. 29, pp. 365–379, Dec. 2015.
S. M. Smith, “Fast robust automated brain extraction,” Human Brain Mapping, vol. 17, pp. 143–155, Nov. 2002.
D. W. Shattuck, S. R. Sandor-Leahy, K. A. Schaper, D. A. Rottenberg, and R. M. Leahy, “Magnetic resonance image tissue classification using a partial volume model,” NeuroImage, vol. 13, pp. 856–876, May 2001.
“Analysis of functional neuroimages.” http://afni.nimh.nih.gov.
A. Mikheev, G. Nevsky, S. Govindan, R. Grossman, and H. Rusinek, “Fully automatic segmentation of the brain from t1-weighted MRI using bridge burner algorithm,” Journal of Magnetic Resonance Imaging, vol. 27, pp. 1235–1241, May 2008.
S. A. Sadananthan, W. Zheng, M. W. Chee, and V. Zagorodnov, “Skull stripping using graph cuts,” NeuroImage, vol. 49, pp. 225–239, Jan. 2010.
F. Ségonne, A. Dale, E. Busa, M. Glessner, D. Salat, H. Hahn, and B. Fischl, “A hybrid approach to the skull stripping problem in mri,” NeuroImage, vol. 22, no. 3, pp. 1060 – 1075, 2004. 15 IEEE LATIN AMERICA TRANSACTIONS , Vol. 7, No. 7, Oct 2020
O. Lucena, R. Souza, L. Rittner, R. Frayne, and R. Lotufo, “Convolutional neural networks for skull-stripping in brain mr imaging using silver standard masks,” Artificial Intelligence in Medicine, vol. 98,
pp. 48–58, 2019.
D. H. M. Nguyen, D. M. Nguyen, M. T. N. Truong, T. Nguyen, K. T. Tran, N. A. Triet, P. T. Bao, and B. T. Nguyen, “Asmcnn: An efficient brain extraction using active shape model and convolutional neural
networks,” 2020.
D. S. Marcus, T. H. Wang, J. Parker, J. G. Csernansky, J. C. Morris, and R. L. Buckner, “Open access series of imaging studies (OASIS): Crosssectional MRI data in young, middle aged, nondemented, and demented older adults,” Journal of Cognitive Neuroscience, vol. 19, pp. 1498–1507, Sept. 2007.
C. B. Barber, D. P. Dobkin, and H. Huhdanpaa, “The quickhull algorithm for convex hulls,” ACM Trans. Math. Softw., vol. 22, p. 469–483, Dec.1996.